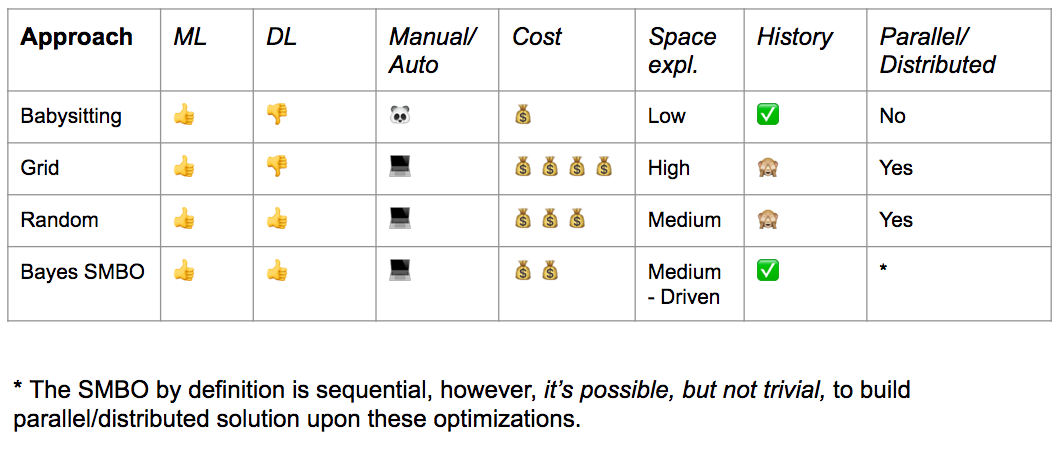
Hyperparameter Optimization Example¶
This Jupyter Notebook is made for illustrating - through a mixture of slides and code in an interactive fashion - the different methods for optimising Hyperparameters for Machine Learning models. First it shows the most naive, manual approach, then grid search, and finally bayesian optimization.
Authors and Date:¶
- Christian Michelsen & Troels Petersen (Niels Bohr Institute)
- 2022-05-01 (latest update)
- Naive, manual approach
- Grid search
- Random search
- Bayesian optimization
- "Full" scan over parameter space
- New methods
- New software
- Focus on the understanding of HPO, not the actual code nor the data!
Nomenclature (i.e. naming scheme)¶
- Machine Learning Model: $\mathcal{A}$
- $N$ hyperparameters
- Domain: $\Lambda_n$
- Hyperparameter configuration space: $\mathbf{\Lambda}=\Lambda_1 \times \Lambda_2 \times \dots \times \Lambda_N $
- Vector of hyperparameters: $\mathbf{\lambda} \in \mathbf{\Lambda}$
- Specific ML model: $\mathcal{A}_\mathbf{\lambda}$
Domain of hyperparameters:¶
- real
- integer
- binary
- categorical
Goal:¶
Given a dataset $\mathcal{D}$, find the vector of HyperParameters $\mathbf{\lambda}^{*}$, which performes "best", i.e. minimises the expected loss function $\mathcal{L}$ for the model $\mathcal{A}_\mathbf{\lambda}$ on the test set of the data $D_\mathrm{test}$:
$$ \mathbf{\lambda}^{*} = \mathop{\mathrm{argmin}}_{\mathbf{\lambda} \in \mathbf{\Lambda}} \mathbb{E}_{D_\mathrm{test} \thicksim \mathcal{D}} \, \left[ \mathbf{V}\left(\mathcal{L}, \mathcal{A}_\mathbf{\lambda}, D_\mathrm{test}\right) \right] $$In practice we have to approximate the expectation above.
First, we import the modules we want to use:
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from sklearn.tree import DecisionTreeClassifier, export_graphviz
from sklearn import tree
from sklearn.datasets import load_iris, load_wine
from sklearn.metrics import accuracy_score
from IPython.display import SVG
from graphviz import Source
from IPython.display import display
from ipywidgets import interactive
from sklearn.model_selection import train_test_split, GridSearchCV, RandomizedSearchCV
from scipy.stats import randint, poisson
import warnings
# warnings.filterwarnings('ignore')
We read in the data:
df = pd.read_csv('./data/Pulsar_data.csv')
df.head(10)
| Mean_SNR | STD_SNR | Kurtosis_SNR | Skewness_SNR | Class | |
|---|---|---|---|---|---|
| 0 | 27.555184 | 61.719016 | 2.208808 | 3.662680 | 1 |
| 1 | 1.358696 | 13.079034 | 13.312141 | 212.597029 | 1 |
| 2 | 73.112876 | 62.070220 | 1.268206 | 1.082920 | 1 |
| 3 | 146.568562 | 82.394624 | -0.274902 | -1.121848 | 1 |
| 4 | 6.071070 | 29.760400 | 5.318767 | 28.698048 | 1 |
| 5 | 32.919732 | 65.094197 | 1.605538 | 0.871364 | 1 |
| 6 | 34.101171 | 62.577395 | 1.890020 | 2.572133 | 1 |
| 7 | 50.107860 | 66.321825 | 1.456423 | 1.335182 | 1 |
| 8 | 176.119565 | 59.737720 | -1.785377 | 2.940913 | 1 |
| 9 | 183.622910 | 79.932815 | -1.326647 | 0.346712 | 1 |
We then divide the dataset in input features (X) and target (y):
X = df.drop(columns='Class')
y = df['Class']
feature_names = df.columns.tolist()[:-1]
print(X.shape)
X_train, X_test, y_train, y_test = train_test_split(X,
y,
test_size=0.20,
random_state=42)
X_train.head(10)
(3278, 4)
| Mean_SNR | STD_SNR | Kurtosis_SNR | Skewness_SNR | |
|---|---|---|---|---|
| 233 | 159.849498 | 76.740010 | -0.575016 | -0.941293 |
| 831 | 4.243311 | 26.746490 | 7.110978 | 52.701218 |
| 2658 | 1.015050 | 10.449662 | 15.593479 | 316.011541 |
| 2495 | 2.235786 | 19.071848 | 9.659137 | 99.294390 |
| 2603 | 2.266722 | 15.512103 | 9.062942 | 99.652157 |
| 111 | 121.404682 | 47.965569 | 0.663053 | 1.203139 |
| 1370 | 35.209866 | 60.573157 | 1.635995 | 1.609377 |
| 1124 | 199.577759 | 58.656643 | -1.862320 | 2.391870 |
| 2170 | 0.663043 | 8.571517 | 23.415092 | 655.614875 |
| 2177 | 3.112876 | 16.855717 | 8.301954 | 90.378150 |
And check out the y values (which turns out to be balanced):
y_train.head(10)
233 1 831 1 2658 0 2495 0 2603 0 111 1 1370 1 1124 1 2170 0 2177 0 Name: Class, dtype: int64
y_train.value_counts()
0 1319 1 1303 Name: Class, dtype: int64
Part A: Naïve Approach¶
- Manual configuration
- "Babysitting is also known as Trial & Error or Grad Student Descent in the academic field"
def fit_and_grapth_estimator(estimator):
estimator.fit(X_train, y_train)
accuracy = accuracy_score(y_train, estimator.predict(X_train))
print(f'Training Accuracy: {accuracy:.4f}')
class_names = [str(i) for i in range(y_train.nunique())]
graph = Source(tree.export_graphviz(estimator,
out_file=None,
feature_names=feature_names,
class_names=class_names,
filled = True))
display(SVG(graph.pipe(format='svg')))
return estimator
def plot_tree(max_depth=1, min_samples_leaf=1):
estimator = DecisionTreeClassifier(random_state=42,
max_depth=max_depth,
min_samples_leaf=min_samples_leaf)
return fit_and_grapth_estimator(estimator)
display(interactive(plot_tree,
max_depth=(1, 10, 1),
min_samples_leaf=(1, 100, 1)))
(Test this interactively in notebook)
And test this configuration out on the test data:
clf_manual = DecisionTreeClassifier(random_state=42,
max_depth=10,
min_samples_leaf=5)
clf_manual.fit(X_train, y_train)
accuracy_manual = accuracy_score(y_test, clf_manual.predict(X_test))
print(f'Accuracy Manual: {accuracy_manual:.4f}')
Accuracy Manual: 0.8201
Part B: Grid Search¶
Grid Search:
- full factorial design
- Cartesian product
- Curse of dimensionality (grows exponentially)
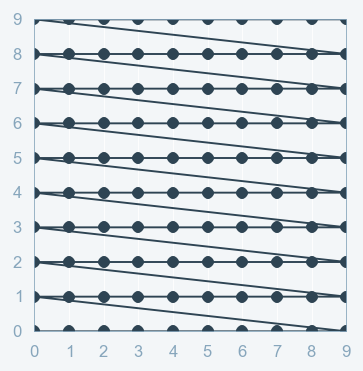
Grid Search with Scikit Learn:
parameters_GridSearch = {'max_depth':[1, 10, 100],
'min_samples_leaf':[1, 10, 100],
}
clf_DecisionTree = DecisionTreeClassifier(random_state=42)
GridSearch = GridSearchCV(clf_DecisionTree,
parameters_GridSearch,
cv=5,
return_train_score=True,
refit=True,
)
GridSearch.fit(X_train, y_train);
GridSearch_results = pd.DataFrame(GridSearch.cv_results_)
print("Grid Search: \tBest parameters: ", GridSearch.best_params_, f", Best scores: {GridSearch.best_score_:.4f}\n")
Grid Search: Best parameters: {'max_depth': 1, 'min_samples_leaf': 1} , Best scores: 0.8551
GridSearch_results
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_max_depth | param_min_samples_leaf | params | split0_test_score | split1_test_score | split2_test_score | ... | mean_test_score | std_test_score | rank_test_score | split0_train_score | split1_train_score | split2_train_score | split3_train_score | split4_train_score | mean_train_score | std_train_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.002718 | 0.000376 | 0.000753 | 0.000065 | 1 | 1 | {'max_depth': 1, 'min_samples_leaf': 1} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 1 | 0.002312 | 0.000131 | 0.000671 | 0.000039 | 1 | 10 | {'max_depth': 1, 'min_samples_leaf': 10} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 2 | 0.001979 | 0.000029 | 0.000585 | 0.000007 | 1 | 100 | {'max_depth': 1, 'min_samples_leaf': 100} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 3 | 0.006214 | 0.000106 | 0.000621 | 0.000002 | 10 | 1 | {'max_depth': 10, 'min_samples_leaf': 1} | 0.847619 | 0.822857 | 0.858779 | ... | 0.841729 | 0.011991 | 8 | 0.956128 | 0.948021 | 0.954242 | 0.962345 | 0.957102 | 0.955568 | 0.004633 |
| 4 | 0.005719 | 0.000062 | 0.000631 | 0.000015 | 10 | 10 | {'max_depth': 10, 'min_samples_leaf': 10} | 0.845714 | 0.847619 | 0.853053 | ... | 0.846300 | 0.008757 | 6 | 0.896042 | 0.898903 | 0.898475 | 0.893232 | 0.895615 | 0.896453 | 0.002066 |
| 5 | 0.003874 | 0.000159 | 0.000615 | 0.000030 | 10 | 100 | {'max_depth': 10, 'min_samples_leaf': 100} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 6 | 0.006463 | 0.000121 | 0.000604 | 0.000025 | 100 | 1 | {'max_depth': 100, 'min_samples_leaf': 1} | 0.826667 | 0.796190 | 0.841603 | ... | 0.824190 | 0.015056 | 9 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 0.000000 |
| 7 | 0.005515 | 0.000063 | 0.000600 | 0.000026 | 100 | 10 | {'max_depth': 100, 'min_samples_leaf': 10} | 0.845714 | 0.849524 | 0.854962 | ... | 0.845154 | 0.009863 | 7 | 0.896996 | 0.899857 | 0.898475 | 0.894185 | 0.896568 | 0.897216 | 0.001909 |
| 8 | 0.003703 | 0.000104 | 0.000630 | 0.000114 | 100 | 100 | {'max_depth': 100, 'min_samples_leaf': 100} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
9 rows × 22 columns
clf_GridSearch = GridSearch.best_estimator_
accuracy_GridSearch = accuracy_score(y_test, clf_GridSearch.predict(X_test))
print(f'Accuracy Manual: {accuracy_manual:.4f}')
print(f'Accuracy Grid Search: {accuracy_GridSearch:.4f}')
Accuracy Manual: 0.8201 Accuracy Grid Search: 0.8430
Part C: Random Search¶
- $B$ function evaluations, $N$ hyperparameters, $y$ number of different values:
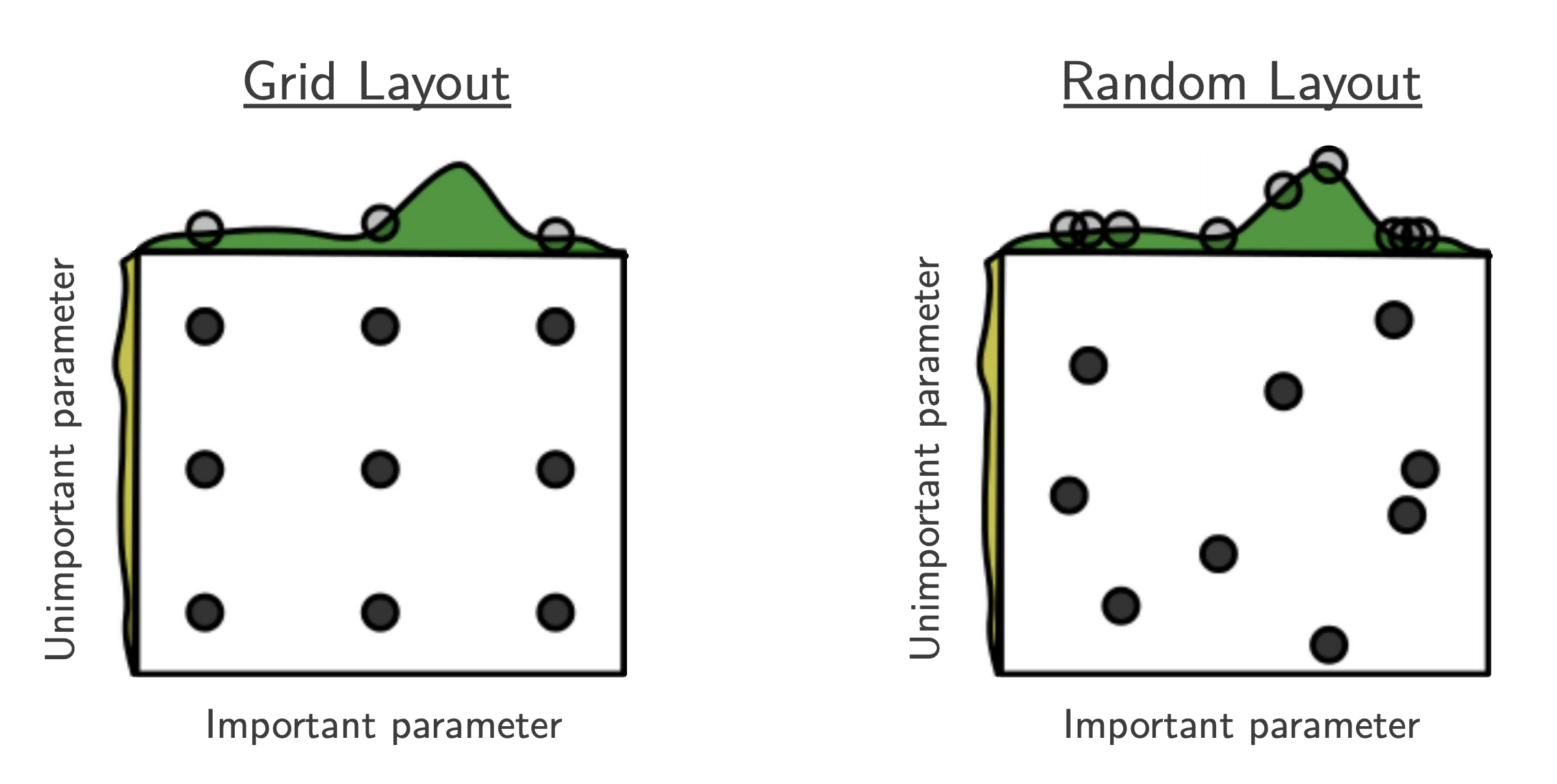
- "This failure of grid search is the rule rather than the exception in high dimensional hyper-parameter optimization" [Bergstra, 2012]
- useful baseline because (almost) no assumptions about the ML algorithm being optimized.
Random Search with Scikit Learn using Scipy Stats as PDFs for the parameters:
# specify parameters and distributions to sample from
parameters_RandomSearch = {'max_depth': poisson(25),
'min_samples_leaf': randint(1, 100)}
# run randomized search
n_iter_search = 9
RandomSearch = RandomizedSearchCV(clf_DecisionTree,
param_distributions=parameters_RandomSearch,
n_iter=n_iter_search,
cv=5,
return_train_score=True,
random_state=42,
)
# fit the random search instance
RandomSearch.fit(X_train, y_train);
RandomSearch_results = pd.DataFrame(RandomSearch.cv_results_)
print("Random Search: \tBest parameters: ", RandomSearch.best_params_, f", Best scores: {RandomSearch.best_score_:.3f}")
Random Search: Best parameters: {'max_depth': 26, 'min_samples_leaf': 83} , Best scores: 0.855
RandomSearch_results.head(10)
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_max_depth | param_min_samples_leaf | params | split0_test_score | split1_test_score | split2_test_score | ... | mean_test_score | std_test_score | rank_test_score | split0_train_score | split1_train_score | split2_train_score | split3_train_score | split4_train_score | mean_train_score | std_train_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.005877 | 0.000319 | 0.000910 | 0.000122 | 23 | 72 | {'max_depth': 23, 'min_samples_leaf': 72} | 0.849524 | 0.862857 | 0.862595 | ... | 0.854308 | 0.010440 | 7 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 1 | 0.004484 | 0.000305 | 0.000655 | 0.000046 | 26 | 83 | {'max_depth': 26, 'min_samples_leaf': 83} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 2 | 0.005210 | 0.000050 | 0.000611 | 0.000002 | 17 | 24 | {'max_depth': 17, 'min_samples_leaf': 24} | 0.847619 | 0.860952 | 0.868321 | ... | 0.854310 | 0.012162 | 6 | 0.875536 | 0.879351 | 0.878456 | 0.875596 | 0.881792 | 0.878146 | 0.002373 |
| 3 | 0.003949 | 0.000080 | 0.000587 | 0.000014 | 27 | 88 | {'max_depth': 27, 'min_samples_leaf': 88} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 4 | 0.004144 | 0.000078 | 0.000598 | 0.000027 | 31 | 64 | {'max_depth': 31, 'min_samples_leaf': 64} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.861296 | 0.860031 | 0.001460 |
| 5 | 0.003952 | 0.000098 | 0.000585 | 0.000005 | 27 | 89 | {'max_depth': 27, 'min_samples_leaf': 89} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 6 | 0.004548 | 0.000031 | 0.000581 | 0.000009 | 22 | 42 | {'max_depth': 22, 'min_samples_leaf': 42} | 0.849524 | 0.836190 | 0.856870 | ... | 0.845540 | 0.015574 | 9 | 0.866476 | 0.865999 | 0.862726 | 0.865110 | 0.864156 | 0.864893 | 0.001343 |
| 7 | 0.004142 | 0.000094 | 0.000568 | 0.000006 | 21 | 62 | {'max_depth': 21, 'min_samples_leaf': 62} | 0.849524 | 0.840000 | 0.862595 | ... | 0.850500 | 0.009778 | 8 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.861296 | 0.860031 | 0.001460 |
| 8 | 0.004051 | 0.000044 | 0.000573 | 0.000016 | 20 | 64 | {'max_depth': 20, 'min_samples_leaf': 64} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.861296 | 0.860031 | 0.001460 |
9 rows × 22 columns
clf_RandomSearch = RandomSearch.best_estimator_
accuracy_RandomSearch = accuracy_score(y_test, clf_RandomSearch.predict(X_test))
print(f'Accuracy Manual: {accuracy_manual:.4f}')
print(f'Accuracy Grid search: {accuracy_GridSearch:.4f}')
print(f'Accuracy Random Search: {accuracy_RandomSearch:.4f}')
Accuracy Manual: 0.8201 Accuracy Grid search: 0.8430 Accuracy Random Search: 0.8430
Part D: Bayesian Optimization¶
- Expensive black box functions $\Rightarrow$ need of smart guesses
- Probabilistic Surrogate Model (to be fitted)
- Often Gaussian Processes
- Acquisition function
- Exploitation / Exploration
- Cheap to Computer
[Brochu, Cora, de Freitas, 2010]
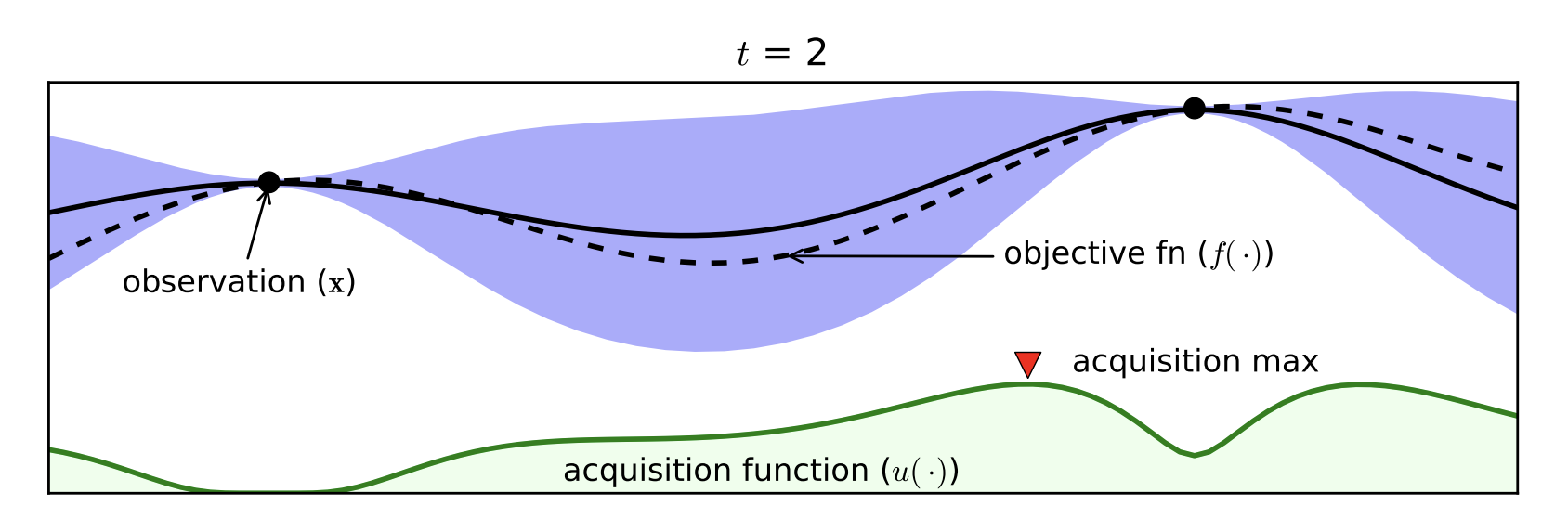
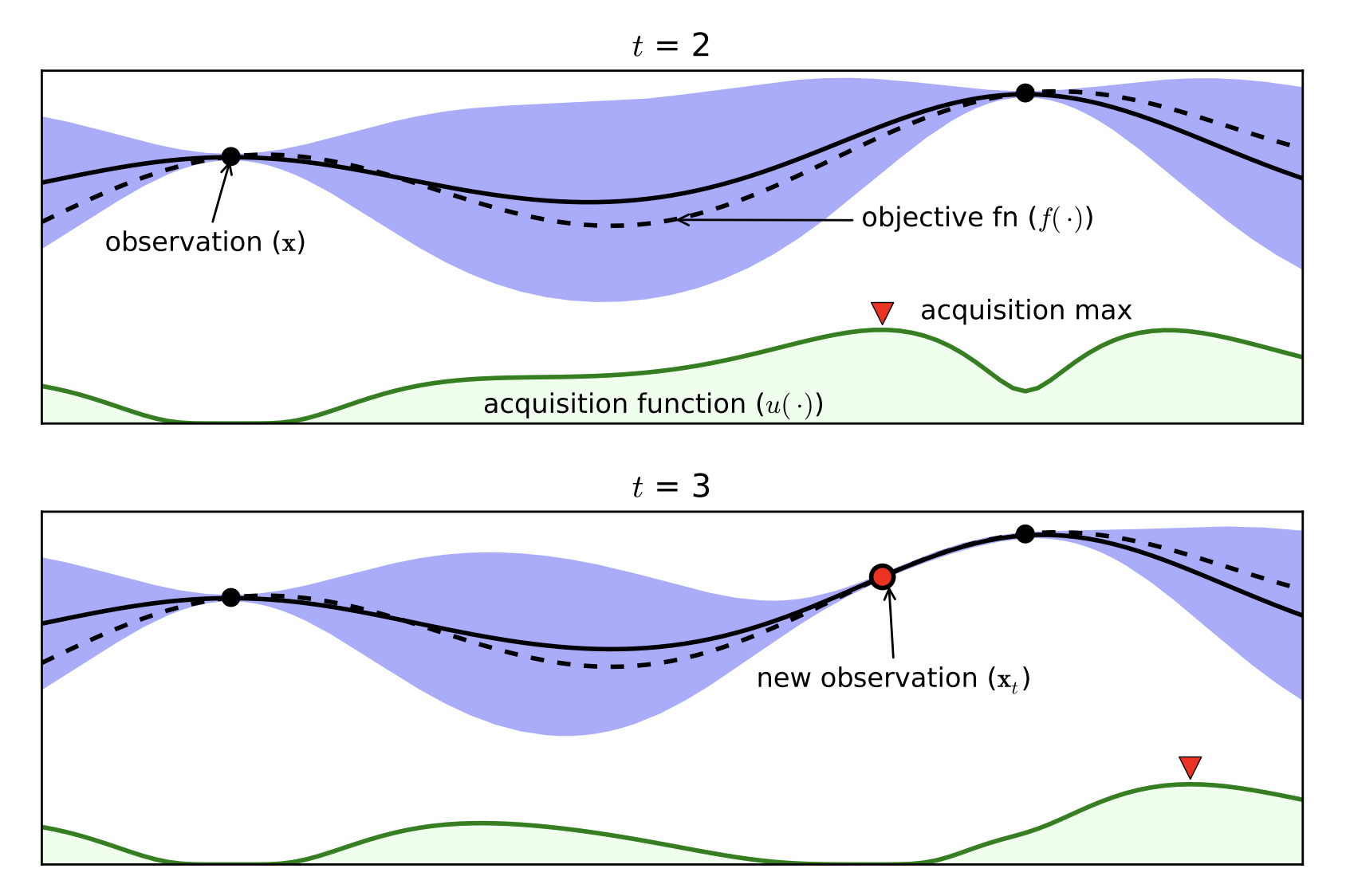
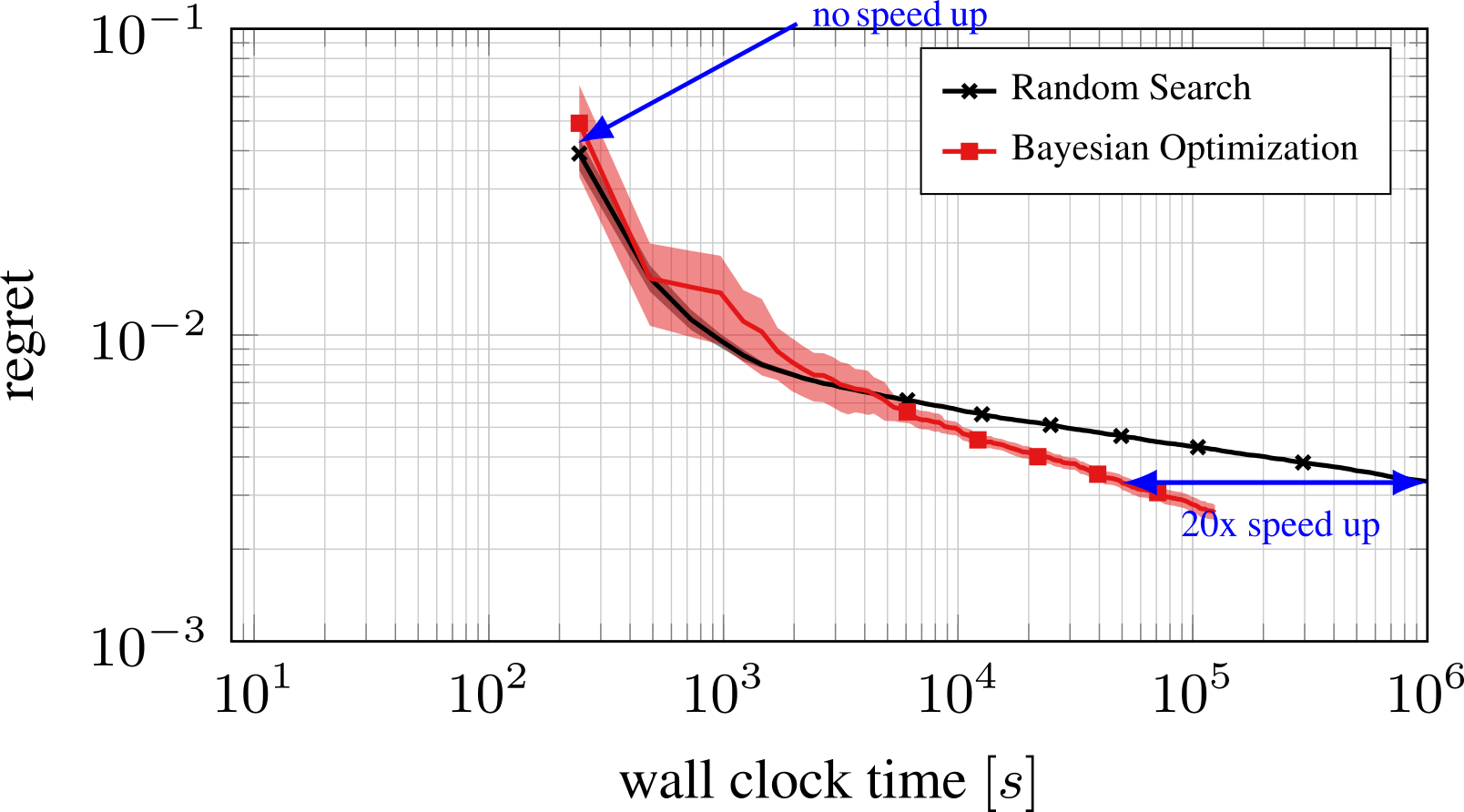
Bayesian Optimization with the Python package BayesianOptimization:
from bayes_opt import BayesianOptimization
from sklearn.model_selection import cross_val_score
def DecisionTree_CrossValidation(max_depth, min_samples_leaf, data, targets):
"""Decision Tree cross validation.
Fits a Decision Tree with the given paramaters to the target
given data, calculated a CV accuracy score and returns the mean.
The goal is to find combinations of max_depth, min_samples_leaf
that maximize the accuracy
"""
estimator = DecisionTreeClassifier(random_state=42,
max_depth=max_depth,
min_samples_leaf=min_samples_leaf)
cval = cross_val_score(estimator, data, targets, scoring='accuracy', cv=5)
return cval.mean()
def optimize_DecisionTree(data, targets, pars, n_iter=5):
"""Apply Bayesian Optimization to Decision Tree parameters."""
def crossval_wrapper(max_depth, min_samples_leaf):
"""Wrapper of Decision Tree cross validation.
Notice how we ensure max_depth, min_samples_leaf
are casted to integer before we pass them along.
"""
return DecisionTree_CrossValidation(max_depth=int(max_depth),
min_samples_leaf=int(min_samples_leaf),
data=data,
targets=targets)
optimizer = BayesianOptimization(f=crossval_wrapper,
pbounds=pars,
random_state=42,
verbose=2)
optimizer.maximize(init_points=4, n_iter=n_iter)
return optimizer
parameters_BayesianOptimization = {"max_depth": (1, 100),
"min_samples_leaf": (1, 100),
}
BayesianOptimization = optimize_DecisionTree(X_train,
y_train,
parameters_BayesianOptimization,
n_iter=5)
print(BayesianOptimization.max)
| iter | target | max_depth | min_sa... |
-------------------------------------------------
| 1 | 0.8551 | 38.08 | 95.12 |
| 2 | 0.849 | 73.47 | 60.27 |
| 3 | 0.8524 | 16.45 | 16.44 |
| 4 | 0.8551 | 6.75 | 86.75 |
| 5 | 0.8551 | 1.56 | 99.21 |
| 6 | 0.8551 | 2.084 | 88.69 |
| 7 | 0.8551 | 1.206 | 97.77 |
| 8 | 0.8551 | 1.452 | 98.8 |
| 9 | 0.8551 | 1.165 | 98.39 |
=================================================
{'target': 0.8550716103235187, 'params': {'max_depth': 38.07947176588889, 'min_samples_leaf': 95.1207163345817}}
params = BayesianOptimization.max['params']
clf_BO = DecisionTreeClassifier(random_state=42, **params)
clf_BO = clf_BO.fit(X_train, y_train)
accuracy_BayesianOptimization = accuracy_score(y_test, clf_BO.predict(X_test))
print(f'Accuracy Manual: {accuracy_manual:.4f}')
print(f'Accuracy Grid Search: {accuracy_GridSearch:.4f}')
print(f'Accuracy Random Search: {accuracy_RandomSearch:.4f}')
print(f'Accuracy Bayesian Optimization: {accuracy_BayesianOptimization:.4f}')
Accuracy Manual: 0.8201 Accuracy Grid Search: 0.8430 Accuracy Random Search: 0.8430 Accuracy Bayesian Optimization: 0.8430
Part D: Full Scan over Parameter Space¶
Only possible in low-dimensional space, slow
max_depth_array = np.arange(1, 30)
min_samples_leaf_array = np.arange(2, 31)
Z = np.zeros((len(max_depth_array), len(min_samples_leaf_array)))
for i, max_depth in enumerate(max_depth_array):
for j, min_samples_leaf in enumerate(min_samples_leaf_array):
clf = DecisionTreeClassifier(random_state=42,
max_depth=max_depth,
min_samples_leaf=
min_samples_leaf)
clf.fit(X_train, y_train)
acc = accuracy_score(y_test, clf.predict(X_test))
Z[i, j] = acc
# Notice: have to transpose Z to match up with imshow
Z = Z.T
Plot the results:
fig, ax = plt.subplots(figsize=(12, 6))
# notice that we are setting the extent and origin keywords
CS = ax.imshow(Z, extent=[1, 30, 2, 31], cmap='viridis', origin='lower')
ax.set(xlabel='max_depth', ylabel='min_samples_leaf')
fig.colorbar(CS);
Sum up:¶
Part E: New Methods¶
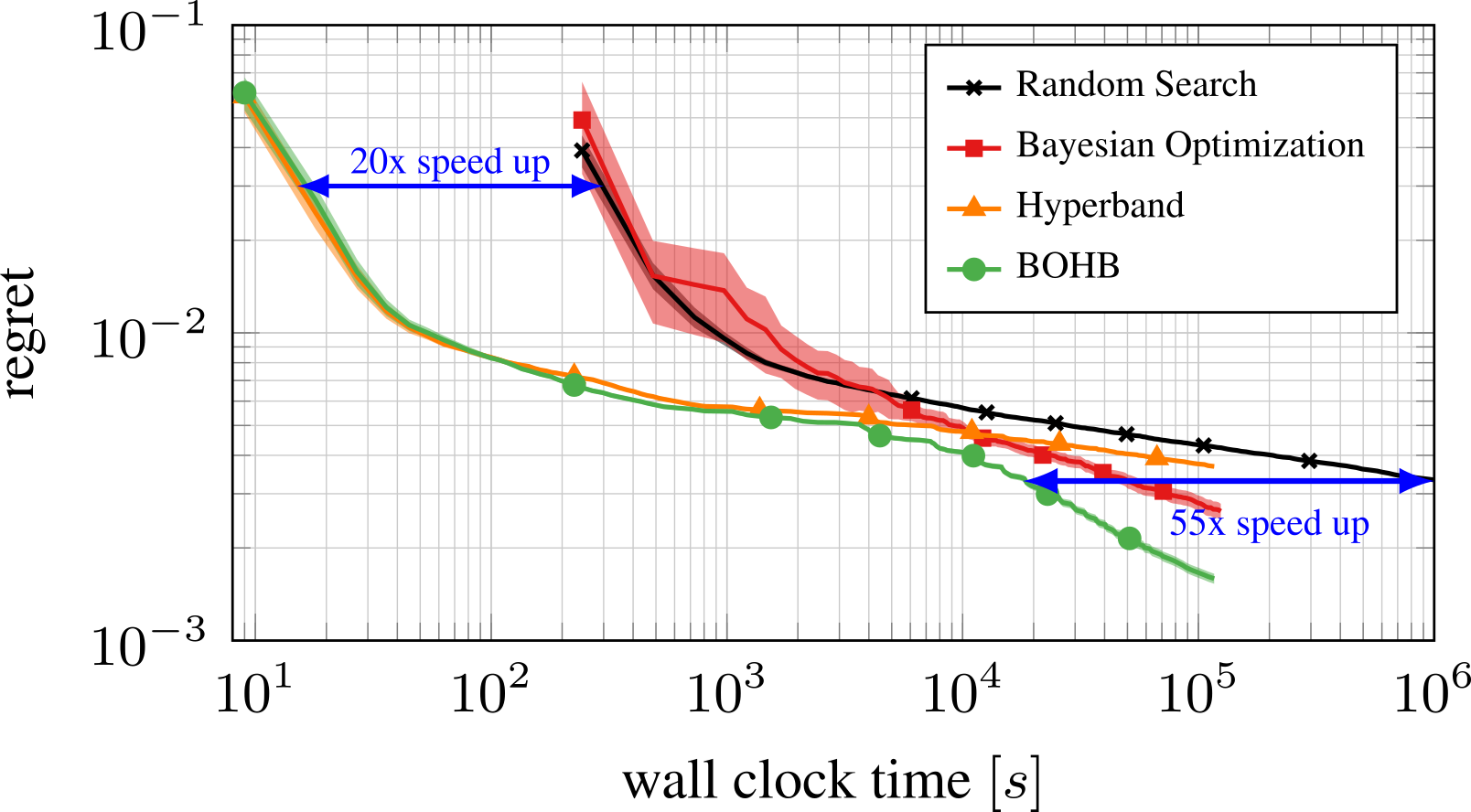
Bayesian Optimization meets HyperBand (BOHB)
HyperBand:
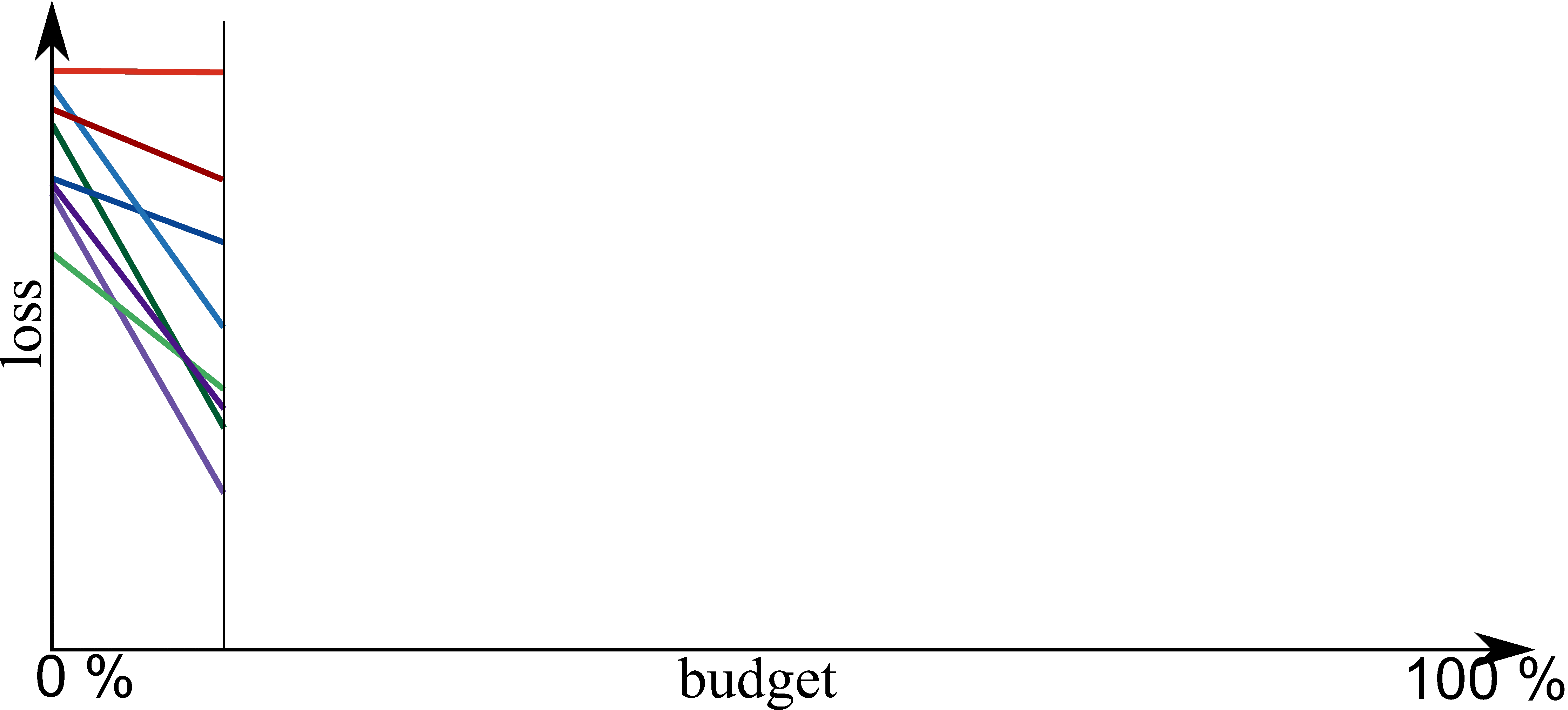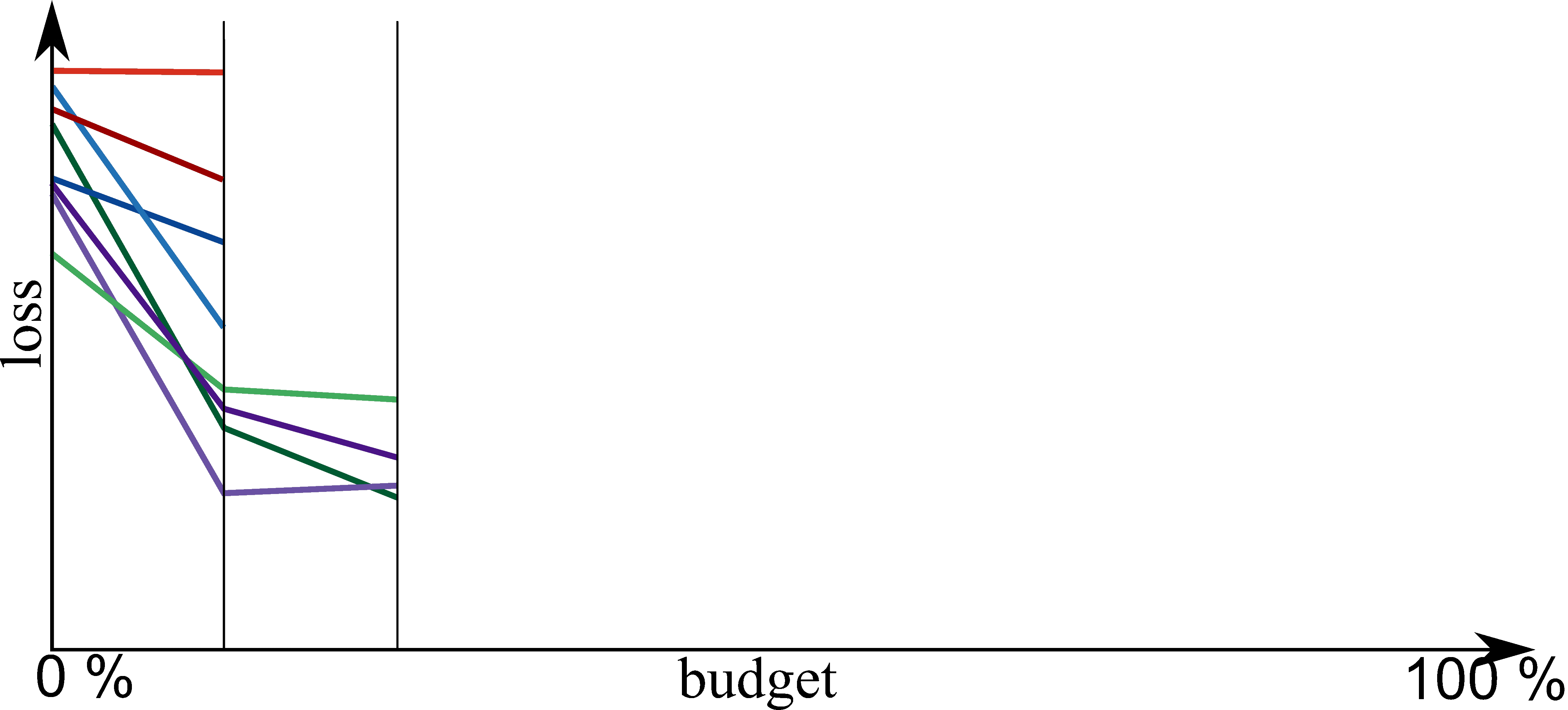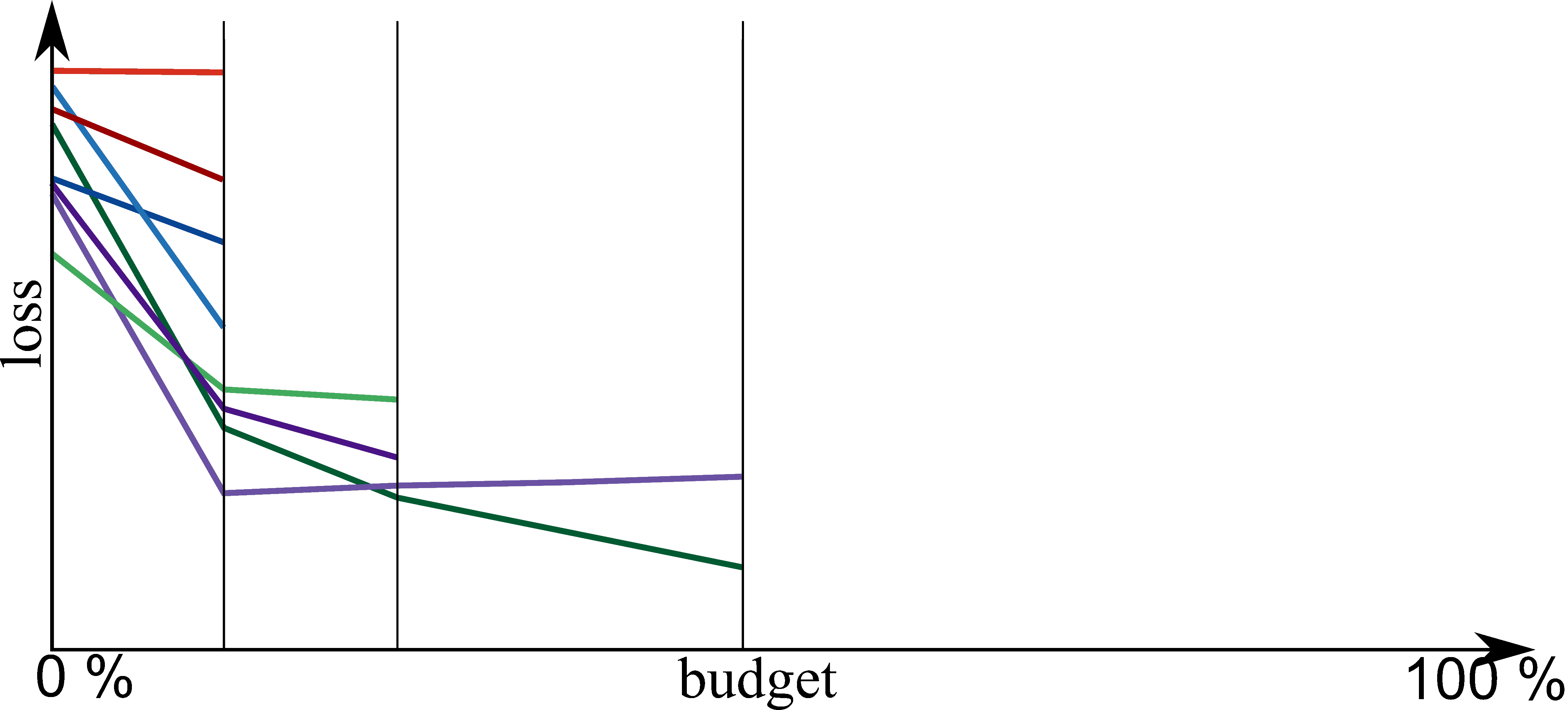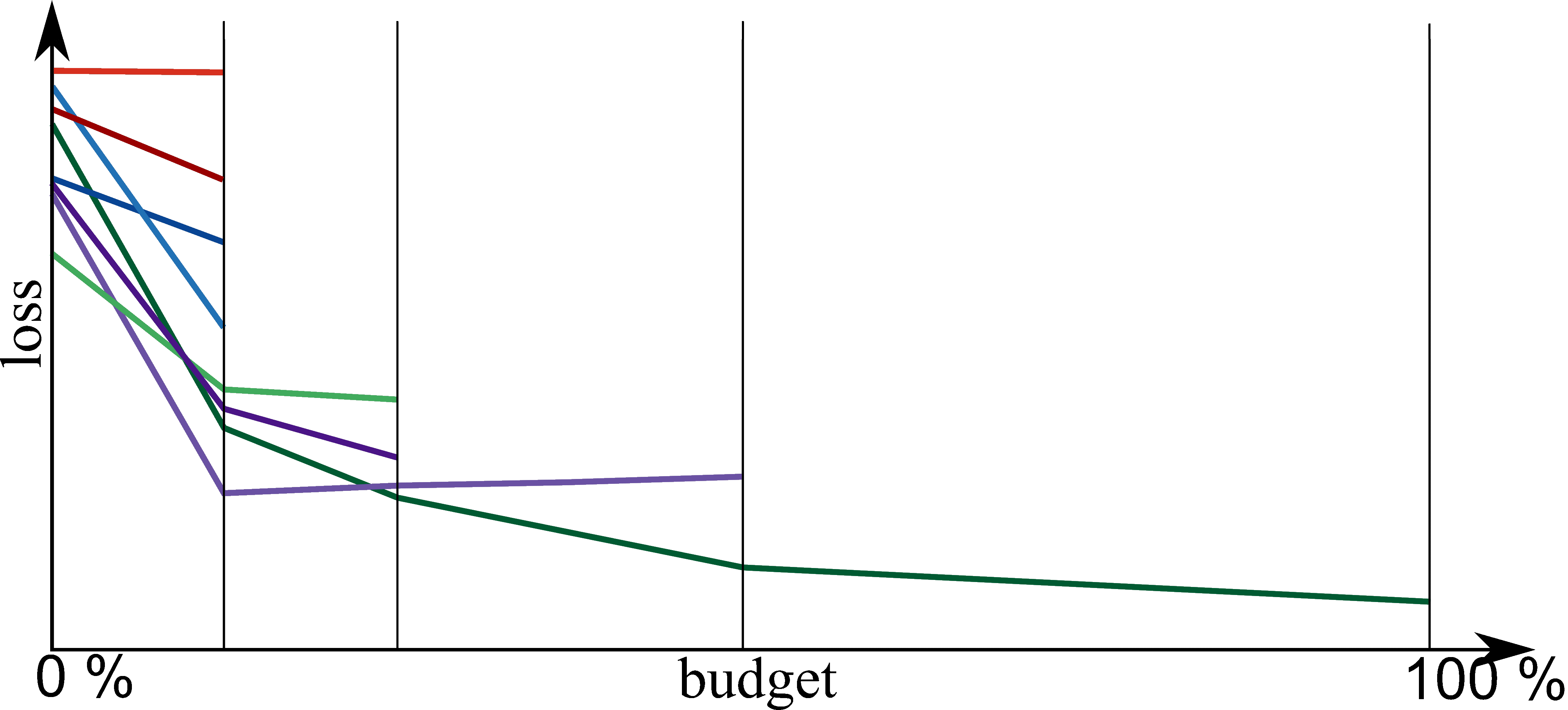

import optuna
from optuna.samplers import TPESampler
from optuna.integration import LightGBMPruningCallback
from optuna.pruners import MedianPruner
import lightgbm as lgb
lgb_data_train = lgb.Dataset(X_train, label=y_train);
/opt/anaconda3/lib/python3.7/site-packages/lightgbm/__init__.py:48: UserWarning: Starting from version 2.2.1, the library file in distribution wheels for macOS is built by the Apple Clang (Xcode_8.3.3) compiler. This means that in case of installing LightGBM from PyPI via the ``pip install lightgbm`` command, you don't need to install the gcc compiler anymore. Instead of that, you need to install the OpenMP library, which is required for running LightGBM on the system with the Apple Clang compiler. You can install the OpenMP library by the following command: ``brew install libomp``. "You can install the OpenMP library by the following command: ``brew install libomp``.", UserWarning)
def objective(trial):
boosting_types = ["gbdt", "rf", "dart"]
boosting_type = trial.suggest_categorical("boosting_type", boosting_types)
params = {
"objective": "binary",
"metric": 'auc',
"boosting": boosting_type,
"max_depth": 5,
"max_depth": trial.suggest_int("max_depth", 2, 63),
"min_child_weight": trial.suggest_loguniform("min_child_weight", 1e-5, 10),
"scale_pos_weight": trial.suggest_uniform("scale_pos_weight", 10.0, 30.0),
"bagging_freq": 1, "bagging_fraction": 0.6,
"verbosity": -1
}
N_iterations_max = 10_000
early_stopping_rounds = 50
if boosting_type == "dart":
N_iterations_max = 100
early_stopping_rounds = None
cv_res = lgb.cv(
params,
lgb_data_train,
num_boost_round=N_iterations_max,
early_stopping_rounds=early_stopping_rounds,
verbose_eval=False,
seed=42,
callbacks=[LightGBMPruningCallback(trial, "auc")],
)
num_boost_round = len(cv_res["auc-mean"])
trial.set_user_attr("num_boost_round", num_boost_round)
return cv_res["auc-mean"][-1]
study = optuna.create_study(
direction="maximize",
sampler=TPESampler(seed=42),
pruner=MedianPruner(n_warmup_steps=50),
)
study.optimize(objective, n_trials=100, show_progress_bar=True);
[I 2022-05-09 14:06:21,594] A new study created in memory with name: no-name-e8a86119-bfbc-476a-b03f-9ebe158adca7
/opt/anaconda3/lib/python3.7/site-packages/optuna/progress_bar.py:47: ExperimentalWarning: Progress bar is experimental (supported from v1.2.0). The interface can change in the future.
self._init_valid()
[I 2022-05-09 14:06:21,898] Trial 0 finished with value: 0.9257274934861961 and parameters: {'boosting_type': 'rf', 'max_depth': 39, 'min_child_weight': 8.632008168602535e-05, 'scale_pos_weight': 13.119890406724053}. Best is trial 0 with value: 0.9257274934861961. [I 2022-05-09 14:06:22,198] Trial 1 finished with value: 0.925645717219157 and parameters: {'boosting_type': 'rf', 'max_depth': 45, 'min_child_weight': 1.3289448722869181e-05, 'scale_pos_weight': 29.398197043239886}. Best is trial 0 with value: 0.9257274934861961. [I 2022-05-09 14:06:22,520] Trial 2 finished with value: 0.9327382743380188 and parameters: {'boosting_type': 'gbdt', 'max_depth': 13, 'min_child_weight': 0.0006690421166498799, 'scale_pos_weight': 20.495128632644757}. Best is trial 2 with value: 0.9327382743380188. [I 2022-05-09 14:06:22,839] Trial 3 finished with value: 0.9293024673628942 and parameters: {'boosting_type': 'dart', 'max_depth': 10, 'min_child_weight': 0.0005660670699258885, 'scale_pos_weight': 17.327236865873836}. Best is trial 2 with value: 0.9327382743380188. [I 2022-05-09 14:06:23,118] Trial 4 finished with value: 0.9261444485440651 and parameters: {'boosting_type': 'rf', 'max_depth': 33, 'min_child_weight': 0.035849855803404704, 'scale_pos_weight': 10.929008254399955}. Best is trial 2 with value: 0.9327382743380188. [I 2022-05-09 14:06:23,491] Trial 5 finished with value: 0.9311118522518811 and parameters: {'boosting_type': 'gbdt', 'max_depth': 60, 'min_child_weight': 6.220025976819156, 'scale_pos_weight': 26.16794696232922}. Best is trial 2 with value: 0.9327382743380188. [I 2022-05-09 14:06:23,640] Trial 6 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:23,840] Trial 7 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:23,979] Trial 8 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:24,121] Trial 9 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:24,178] Trial 10 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:24,505] Trial 11 finished with value: 0.9309290578399502 and parameters: {'boosting_type': 'gbdt', 'max_depth': 63, 'min_child_weight': 6.894420684704665, 'scale_pos_weight': 25.370137676514304}. Best is trial 2 with value: 0.9327382743380188. [I 2022-05-09 14:06:24,849] Trial 12 finished with value: 0.9323419999728675 and parameters: {'boosting_type': 'gbdt', 'max_depth': 63, 'min_child_weight': 0.38601839366790247, 'scale_pos_weight': 23.675597363736365}. Best is trial 2 with value: 0.9327382743380188. [I 2022-05-09 14:06:25,206] Trial 13 finished with value: 0.9327630495788028 and parameters: {'boosting_type': 'gbdt', 'max_depth': 21, 'min_child_weight': 0.47333235522568584, 'scale_pos_weight': 22.116723260828707}. Best is trial 13 with value: 0.9327630495788028. [I 2022-05-09 14:06:25,582] Trial 14 finished with value: 0.9318985480712734 and parameters: {'boosting_type': 'gbdt', 'max_depth': 20, 'min_child_weight': 0.0018017260723627204, 'scale_pos_weight': 20.87074372077165}. Best is trial 13 with value: 0.9327630495788028. [I 2022-05-09 14:06:25,997] Trial 15 finished with value: 0.933907588579396 and parameters: {'boosting_type': 'gbdt', 'max_depth': 23, 'min_child_weight': 0.5262050790257533, 'scale_pos_weight': 17.101924194245463}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:26,403] Trial 16 finished with value: 0.9323761434494685 and parameters: {'boosting_type': 'gbdt', 'max_depth': 25, 'min_child_weight': 0.7139629318981857, 'scale_pos_weight': 15.599819505406096}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:26,506] Trial 17 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:26,918] Trial 18 finished with value: 0.9320047563876231 and parameters: {'boosting_type': 'gbdt', 'max_depth': 20, 'min_child_weight': 0.07473651026301241, 'scale_pos_weight': 18.745829052448148}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:27,094] Trial 19 pruned. Trial was pruned at iteration 61. [I 2022-05-09 14:06:27,515] Trial 20 finished with value: 0.9328372233375839 and parameters: {'boosting_type': 'gbdt', 'max_depth': 21, 'min_child_weight': 1.5423777057795445, 'scale_pos_weight': 13.233732216784787}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:27,931] Trial 21 finished with value: 0.9321245240568861 and parameters: {'boosting_type': 'gbdt', 'max_depth': 22, 'min_child_weight': 1.5417332097862932, 'scale_pos_weight': 10.107893557559724}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:28,270] Trial 22 finished with value: 0.9323386454194766 and parameters: {'boosting_type': 'gbdt', 'max_depth': 26, 'min_child_weight': 0.13230118984954353, 'scale_pos_weight': 12.488404178785878}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:28,440] Trial 23 pruned. Trial was pruned at iteration 66. [I 2022-05-09 14:06:28,827] Trial 24 finished with value: 0.9324599798628743 and parameters: {'boosting_type': 'gbdt', 'max_depth': 29, 'min_child_weight': 0.22938248553821958, 'scale_pos_weight': 18.52693332188188}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:29,148] Trial 25 finished with value: 0.9323709490119938 and parameters: {'boosting_type': 'gbdt', 'max_depth': 10, 'min_child_weight': 0.014223320052048582, 'scale_pos_weight': 11.577261921985883}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:29,293] Trial 26 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:29,623] Trial 27 finished with value: 0.932944700613427 and parameters: {'boosting_type': 'gbdt', 'max_depth': 37, 'min_child_weight': 0.018459882906772026, 'scale_pos_weight': 17.679098751711575}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:29,986] Trial 28 finished with value: 0.9327459260646458 and parameters: {'boosting_type': 'gbdt', 'max_depth': 38, 'min_child_weight': 0.006874274339719773, 'scale_pos_weight': 14.404674789343591}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:30,123] Trial 29 pruned. Trial was pruned at iteration 52. [I 2022-05-09 14:06:30,512] Trial 30 finished with value: 0.9327176880494863 and parameters: {'boosting_type': 'gbdt', 'max_depth': 51, 'min_child_weight': 0.003257102075046086, 'scale_pos_weight': 17.435919490699508}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:30,641] Trial 31 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:30,789] Trial 32 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:31,139] Trial 33 finished with value: 0.9326676138454196 and parameters: {'boosting_type': 'gbdt', 'max_depth': 23, 'min_child_weight': 2.9350751934489323, 'scale_pos_weight': 17.738460794183766}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:31,270] Trial 34 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:31,414] Trial 35 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:31,541] Trial 36 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:31,697] Trial 37 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:31,824] Trial 38 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:32,201] Trial 39 finished with value: 0.9331977959415845 and parameters: {'boosting_type': 'gbdt', 'max_depth': 35, 'min_child_weight': 0.000341792895071912, 'scale_pos_weight': 10.091883058577956}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:32,345] Trial 40 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:32,711] Trial 41 finished with value: 0.9330434512538387 and parameters: {'boosting_type': 'gbdt', 'max_depth': 36, 'min_child_weight': 0.0010904325188218595, 'scale_pos_weight': 10.103269089092729}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:33,068] Trial 42 finished with value: 0.9330358105424482 and parameters: {'boosting_type': 'gbdt', 'max_depth': 35, 'min_child_weight': 0.0004148881183051914, 'scale_pos_weight': 10.017562759585447}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:33,217] Trial 43 pruned. Trial was pruned at iteration 56. [I 2022-05-09 14:06:33,367] Trial 44 pruned. Trial was pruned at iteration 51. [I 2022-05-09 14:06:33,723] Trial 45 finished with value: 0.9329793149860837 and parameters: {'boosting_type': 'gbdt', 'max_depth': 41, 'min_child_weight': 0.001167290226730846, 'scale_pos_weight': 10.044747367447195}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:33,871] Trial 46 pruned. Trial was pruned at iteration 56. [I 2022-05-09 14:06:34,027] Trial 47 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:34,156] Trial 48 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:34,507] Trial 49 finished with value: 0.9331977959415845 and parameters: {'boosting_type': 'gbdt', 'max_depth': 31, 'min_child_weight': 0.0029537732097114785, 'scale_pos_weight': 10.091379098007323}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:34,847] Trial 50 finished with value: 0.9332437207336415 and parameters: {'boosting_type': 'gbdt', 'max_depth': 31, 'min_child_weight': 0.0036815043901656367, 'scale_pos_weight': 11.101495735354064}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:35,065] Trial 51 pruned. Trial was pruned at iteration 80. [I 2022-05-09 14:06:35,196] Trial 52 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:35,337] Trial 53 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:35,682] Trial 54 finished with value: 0.9326659755677553 and parameters: {'boosting_type': 'gbdt', 'max_depth': 32, 'min_child_weight': 0.005148615159009996, 'scale_pos_weight': 10.013233337332471}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:36,029] Trial 55 finished with value: 0.932887914967943 and parameters: {'boosting_type': 'gbdt', 'max_depth': 35, 'min_child_weight': 0.0007769349556585813, 'scale_pos_weight': 10.919188105436339}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:36,158] Trial 56 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:36,299] Trial 57 pruned. Trial was pruned at iteration 51. [I 2022-05-09 14:06:36,429] Trial 58 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:36,579] Trial 59 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:36,926] Trial 60 finished with value: 0.9326659755677553 and parameters: {'boosting_type': 'gbdt', 'max_depth': 32, 'min_child_weight': 0.0125312356630232, 'scale_pos_weight': 10.012683183019575}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:37,132] Trial 61 pruned. Trial was pruned at iteration 78. [I 2022-05-09 14:06:37,267] Trial 62 pruned. Trial was pruned at iteration 51. [I 2022-05-09 14:06:37,614] Trial 63 finished with value: 0.9337278447512649 and parameters: {'boosting_type': 'gbdt', 'max_depth': 43, 'min_child_weight': 0.0025822330028222257, 'scale_pos_weight': 10.12268219759839}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:37,746] Trial 64 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:37,888] Trial 65 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:38,023] Trial 66 pruned. Trial was pruned at iteration 51. [I 2022-05-09 14:06:38,163] Trial 67 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:38,292] Trial 68 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:38,428] Trial 69 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:38,558] Trial 70 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:38,698] Trial 71 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:39,046] Trial 72 finished with value: 0.9330532260771051 and parameters: {'boosting_type': 'gbdt', 'max_depth': 41, 'min_child_weight': 0.0010124114585983697, 'scale_pos_weight': 10.019540916241073}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:39,246] Trial 73 pruned. Trial was pruned at iteration 78. [I 2022-05-09 14:06:39,383] Trial 74 pruned. Trial was pruned at iteration 51. [I 2022-05-09 14:06:39,518] Trial 75 pruned. Trial was pruned at iteration 51. [I 2022-05-09 14:06:39,666] Trial 76 pruned. Trial was pruned at iteration 52. [I 2022-05-09 14:06:39,803] Trial 77 pruned. Trial was pruned at iteration 51. [I 2022-05-09 14:06:40,151] Trial 78 finished with value: 0.9332219468158403 and parameters: {'boosting_type': 'gbdt', 'max_depth': 48, 'min_child_weight': 0.00021426558938719107, 'scale_pos_weight': 10.049409464075314}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:40,281] Trial 79 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:40,432] Trial 80 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:40,779] Trial 81 finished with value: 0.9335821924646988 and parameters: {'boosting_type': 'gbdt', 'max_depth': 40, 'min_child_weight': 8.589036966510786e-05, 'scale_pos_weight': 10.070391130777391}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:40,912] Trial 82 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:41,062] Trial 83 pruned. Trial was pruned at iteration 53. [I 2022-05-09 14:06:41,448] Trial 84 finished with value: 0.9333552874637998 and parameters: {'boosting_type': 'gbdt', 'max_depth': 45, 'min_child_weight': 2.4458989204236287e-05, 'scale_pos_weight': 11.595407882622062}. Best is trial 15 with value: 0.933907588579396. [I 2022-05-09 14:06:41,578] Trial 85 pruned. Trial was pruned at iteration 51. [I 2022-05-09 14:06:41,787] Trial 86 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:41,921] Trial 87 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:42,067] Trial 88 pruned. Trial was pruned at iteration 51. [I 2022-05-09 14:06:42,199] Trial 89 pruned. Trial was pruned at iteration 50. [I 2022-05-09 14:06:42,342] Trial 90 pruned. Trial was pruned at iteration 50.
# To see all info at the best trial use:
study.best_trial
# To print metric values for all trials:
study.best_trial.intermediate_values
# To see distributions from which optuna samples parameters:
study.best_trial.distributions
# To simply get the optimized parameters:
study.best_trial.params
Happy HyperParameter Optimisation!¶
...and remember, that this is useful but not essential in this course.