Hyperparameter Optimization Example¶
This Jupyter Notebook is made for illustrating - through a mixture of slides and code in an interactive fashion - the different methods for optimising Hyperparameters for Machine Learning models. First it shows the most naive, manual approach, then grid search, and finally bayesian optimization.
Authors and Date:¶
- Christian Michelsen & Troels Petersen (Niels Bohr Institute)
- 2025-04-27 (latest update)
- Naive, manual approach
- Grid search
- Random search
- Bayesian optimization
- "Full" scan over parameter space
- New methods
- New software
- Focus on the understanding of HPO, not the actual code nor the data!
Nomenclature (i.e. naming scheme)¶
- Machine Learning Model: $\mathcal{A}$
- $N$ hyperparameters
- Domain: $\Lambda_n$
- Hyperparameter configuration space: $\mathbf{\Lambda}=\Lambda_1 \times \Lambda_2 \times \dots \times \Lambda_N $
- Vector of hyperparameters: $\mathbf{\lambda} \in \mathbf{\Lambda}$
- Specific ML model: $\mathcal{A}_\mathbf{\lambda}$
Domain of hyperparameters:¶
- real
- integer
- binary
- categorical
Goal:¶
Given a dataset $\mathcal{D}$, find the vector of HyperParameters $\mathbf{\lambda}^{*}$, which performes "best", i.e. minimises the expected loss function $\mathcal{L}$ for the model $\mathcal{A}_\mathbf{\lambda}$ on the test set of the data $D_\mathrm{test}$:
$$ \mathbf{\lambda}^{*} = \mathop{\mathrm{argmin}}_{\mathbf{\lambda} \in \mathbf{\Lambda}} \mathbb{E}_{D_\mathrm{test} \thicksim \mathcal{D}} \, \left[ \mathbf{V}\left(\mathcal{L}, \mathcal{A}_\mathbf{\lambda}, D_\mathrm{test}\right) \right] $$
In practice we have to approximate the expectation above.
First, we import the modules we want to use:
Important¶
Make sure that you have some additional packages installed:
pip install graphviz
conda install -c conda-forge bayesian-optimization
conda install -c conda-forge optuna
pip install optuna-integration
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from sklearn.tree import DecisionTreeClassifier, export_graphviz
from sklearn import tree
from sklearn.datasets import load_iris, load_wine
from sklearn.metrics import accuracy_score
from IPython.display import SVG
from graphviz import Source
from IPython.display import display
from ipywidgets import interactive
from sklearn.model_selection import train_test_split, GridSearchCV, RandomizedSearchCV
from scipy.stats import randint, poisson
import warnings
# warnings.filterwarnings('ignore')
We read in the data:
df = pd.read_csv('./data/Pulsar_data.csv')
df.head(10)
| Mean_SNR | STD_SNR | Kurtosis_SNR | Skewness_SNR | Class | |
|---|---|---|---|---|---|
| 0 | 27.555184 | 61.719016 | 2.208808 | 3.662680 | 1 |
| 1 | 1.358696 | 13.079034 | 13.312141 | 212.597029 | 1 |
| 2 | 73.112876 | 62.070220 | 1.268206 | 1.082920 | 1 |
| 3 | 146.568562 | 82.394624 | -0.274902 | -1.121848 | 1 |
| 4 | 6.071070 | 29.760400 | 5.318767 | 28.698048 | 1 |
| 5 | 32.919732 | 65.094197 | 1.605538 | 0.871364 | 1 |
| 6 | 34.101171 | 62.577395 | 1.890020 | 2.572133 | 1 |
| 7 | 50.107860 | 66.321825 | 1.456423 | 1.335182 | 1 |
| 8 | 176.119565 | 59.737720 | -1.785377 | 2.940913 | 1 |
| 9 | 183.622910 | 79.932815 | -1.326647 | 0.346712 | 1 |
We then divide the dataset in input features (X) and target (y):
X = df.drop(columns='Class')
y = df['Class']
feature_names = df.columns.tolist()[:-1]
print(X.shape)
X_train, X_test, y_train, y_test = train_test_split(X,
y,
test_size=0.20,
random_state=42)
X_train.head(10)
(3278, 4)
| Mean_SNR | STD_SNR | Kurtosis_SNR | Skewness_SNR | |
|---|---|---|---|---|
| 233 | 159.849498 | 76.740010 | -0.575016 | -0.941293 |
| 831 | 4.243311 | 26.746490 | 7.110978 | 52.701218 |
| 2658 | 1.015050 | 10.449662 | 15.593479 | 316.011541 |
| 2495 | 2.235786 | 19.071848 | 9.659137 | 99.294390 |
| 2603 | 2.266722 | 15.512103 | 9.062942 | 99.652157 |
| 111 | 121.404682 | 47.965569 | 0.663053 | 1.203139 |
| 1370 | 35.209866 | 60.573157 | 1.635995 | 1.609377 |
| 1124 | 199.577759 | 58.656643 | -1.862320 | 2.391870 |
| 2170 | 0.663043 | 8.571517 | 23.415092 | 655.614875 |
| 2177 | 3.112876 | 16.855717 | 8.301954 | 90.378150 |
And check out the y values (which turns out to be balanced):
y_train.head(10)
233 1 831 1 2658 0 2495 0 2603 0 111 1 1370 1 1124 1 2170 0 2177 0 Name: Class, dtype: int64
y_train.value_counts()
Class 0 1319 1 1303 Name: count, dtype: int64
Part A: Naïve Approach¶
- Manual configuration
- "Babysitting is also known as Trial & Error or Grad Student Descent in the academic field"
def fit_and_grapth_estimator(estimator):
estimator.fit(X_train, y_train)
accuracy = accuracy_score(y_train, estimator.predict(X_train))
print(f'Training Accuracy: {accuracy:.4f}')
class_names = [str(i) for i in range(y_train.nunique())]
graph = Source(tree.export_graphviz(estimator,
out_file=None,
feature_names=feature_names,
class_names=class_names,
filled = True))
display(SVG(graph.pipe(format='svg')))
return estimator
def plot_tree(max_depth=1, min_samples_leaf=1):
estimator = DecisionTreeClassifier(random_state=42,
max_depth=max_depth,
min_samples_leaf=min_samples_leaf)
return fit_and_grapth_estimator(estimator)
display(interactive(plot_tree,
max_depth=(1, 10, 1),
min_samples_leaf=(1, 100, 1)))
interactive(children=(IntSlider(value=1, description='max_depth', max=10, min=1), IntSlider(value=1, descripti…
(Test this interactively in notebook)
And test this configuration out on the test data:
clf_manual = DecisionTreeClassifier(random_state=42,
max_depth=10,
min_samples_leaf=5)
clf_manual.fit(X_train, y_train)
accuracy_manual = accuracy_score(y_test, clf_manual.predict(X_test))
print(f'Accuracy Manual: {accuracy_manual:.4f}')
Accuracy Manual: 0.8201
Part B: Grid Search¶
Grid Search:
- full factorial design
- Cartesian product
- Curse of dimensionality (grows exponentially)
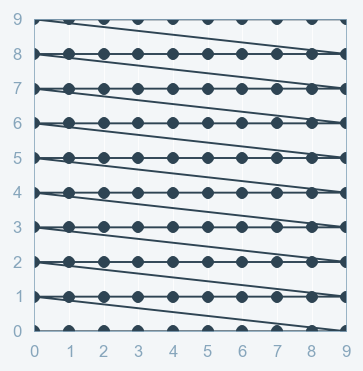
Grid Search with Scikit Learn:
parameters_GridSearch = {'max_depth':[1, 10, 100],
'min_samples_leaf':[1, 10, 100],
}
clf_DecisionTree = DecisionTreeClassifier(random_state=42)
GridSearch = GridSearchCV(clf_DecisionTree,
parameters_GridSearch,
cv=5,
return_train_score=True,
refit=True,
)
GridSearch.fit(X_train, y_train);
GridSearch_results = pd.DataFrame(GridSearch.cv_results_)
print("Grid Search: \tBest parameters: ", GridSearch.best_params_, f", Best scores: {GridSearch.best_score_:.4f}\n")
Grid Search: Best parameters: {'max_depth': 1, 'min_samples_leaf': 1} , Best scores: 0.8551
GridSearch_results
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_max_depth | param_min_samples_leaf | params | split0_test_score | split1_test_score | split2_test_score | ... | mean_test_score | std_test_score | rank_test_score | split0_train_score | split1_train_score | split2_train_score | split3_train_score | split4_train_score | mean_train_score | std_train_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.001210 | 0.000181 | 0.000396 | 0.000039 | 1 | 1 | {'max_depth': 1, 'min_samples_leaf': 1} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 1 | 0.001095 | 0.000055 | 0.000378 | 0.000043 | 1 | 10 | {'max_depth': 1, 'min_samples_leaf': 10} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 2 | 0.001088 | 0.000031 | 0.000371 | 0.000008 | 1 | 100 | {'max_depth': 1, 'min_samples_leaf': 100} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 3 | 0.004697 | 0.000036 | 0.000442 | 0.000051 | 10 | 1 | {'max_depth': 10, 'min_samples_leaf': 1} | 0.847619 | 0.822857 | 0.858779 | ... | 0.841729 | 0.011991 | 8 | 0.956128 | 0.948021 | 0.954242 | 0.962345 | 0.957102 | 0.955568 | 0.004633 |
| 4 | 0.004116 | 0.000087 | 0.000398 | 0.000019 | 10 | 10 | {'max_depth': 10, 'min_samples_leaf': 10} | 0.845714 | 0.847619 | 0.853053 | ... | 0.846682 | 0.008059 | 6 | 0.896042 | 0.898903 | 0.898475 | 0.893232 | 0.895615 | 0.896453 | 0.002066 |
| 5 | 0.002634 | 0.000085 | 0.000392 | 0.000033 | 10 | 100 | {'max_depth': 10, 'min_samples_leaf': 100} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 6 | 0.005000 | 0.000075 | 0.000390 | 0.000011 | 100 | 1 | {'max_depth': 100, 'min_samples_leaf': 1} | 0.826667 | 0.796190 | 0.841603 | ... | 0.824190 | 0.015056 | 9 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 0.000000 |
| 7 | 0.004211 | 0.000122 | 0.000385 | 0.000010 | 100 | 10 | {'max_depth': 100, 'min_samples_leaf': 10} | 0.845714 | 0.849524 | 0.854962 | ... | 0.845536 | 0.009138 | 7 | 0.896996 | 0.899857 | 0.898475 | 0.894185 | 0.896568 | 0.897216 | 0.001909 |
| 8 | 0.002628 | 0.000108 | 0.000371 | 0.000006 | 100 | 100 | {'max_depth': 100, 'min_samples_leaf': 100} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
9 rows × 22 columns
clf_GridSearch = GridSearch.best_estimator_
accuracy_GridSearch = accuracy_score(y_test, clf_GridSearch.predict(X_test))
print(f'Accuracy Manual: {accuracy_manual:.4f}')
print(f'Accuracy Grid Search: {accuracy_GridSearch:.4f}')
Accuracy Manual: 0.8201 Accuracy Grid Search: 0.8430
Part C: Random Search¶
- $B$ function evaluations, $N$ hyperparameters, $y$ number of different values:
$$ y_{\mathrm{Grid Search}} = B^{1/N}, \quad y_{\mathrm{Random Search}} = B $$
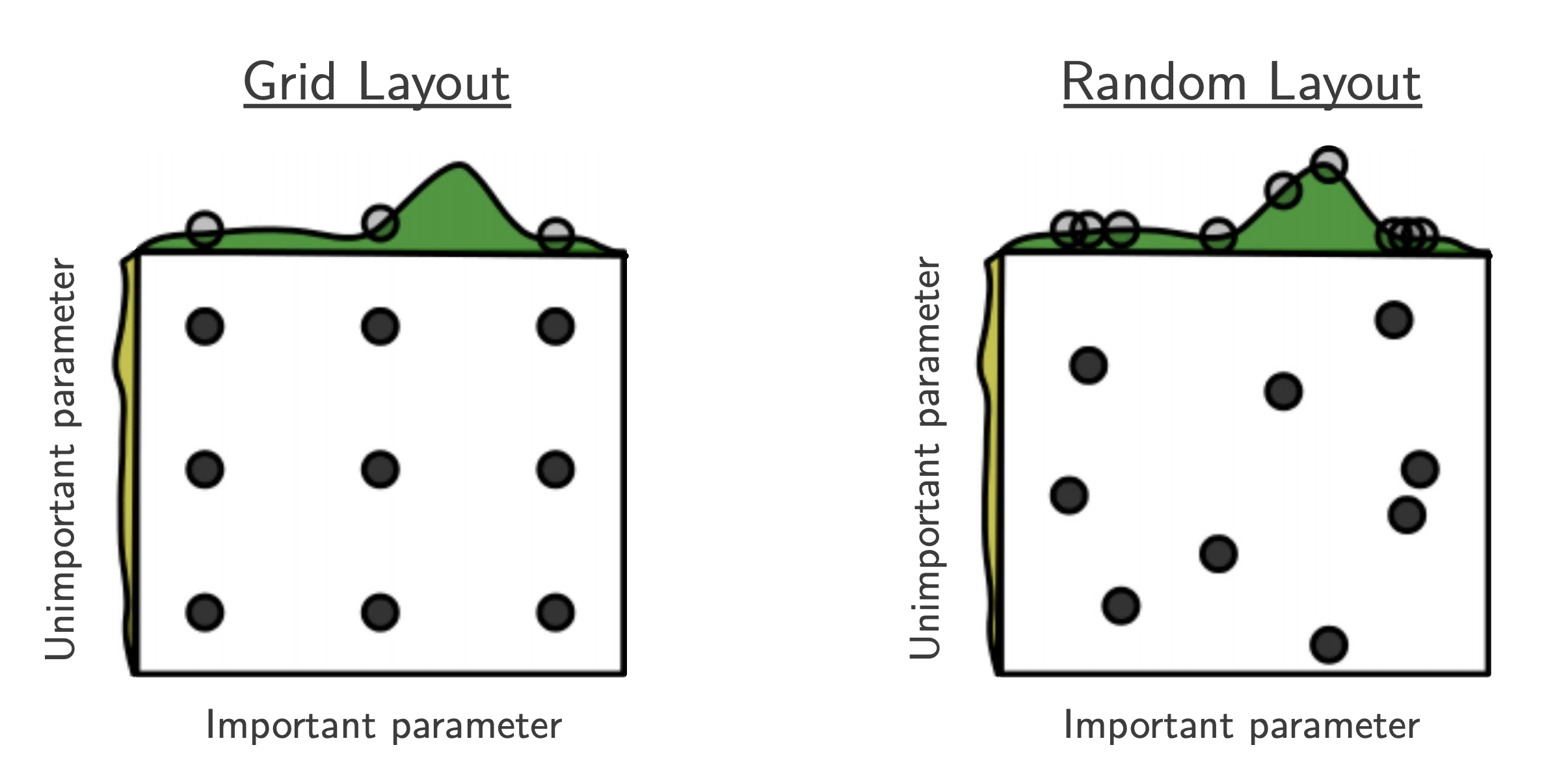
- "This failure of grid search is the rule rather than the exception in high dimensional hyper-parameter optimization" [Bergstra, 2012]
- useful baseline because (almost) no assumptions about the ML algorithm being optimized.
Random Search with Scikit Learn using Scipy Stats as PDFs for the parameters:
# specify parameters and distributions to sample from
parameters_RandomSearch = {'max_depth': poisson(25),
'min_samples_leaf': randint(1, 100)}
# run randomized search
n_iter_search = 9
RandomSearch = RandomizedSearchCV(clf_DecisionTree,
param_distributions=parameters_RandomSearch,
n_iter=n_iter_search,
cv=5,
return_train_score=True,
random_state=42,
)
# fit the random search instance
RandomSearch.fit(X_train, y_train);
RandomSearch_results = pd.DataFrame(RandomSearch.cv_results_)
print("Random Search: \tBest parameters: ", RandomSearch.best_params_, f", Best scores: {RandomSearch.best_score_:.3f}")
Random Search: Best parameters: {'max_depth': 26, 'min_samples_leaf': 83} , Best scores: 0.855
RandomSearch_results.head(10)
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_max_depth | param_min_samples_leaf | params | split0_test_score | split1_test_score | split2_test_score | ... | mean_test_score | std_test_score | rank_test_score | split0_train_score | split1_train_score | split2_train_score | split3_train_score | split4_train_score | mean_train_score | std_train_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.003145 | 0.000194 | 0.000470 | 0.000064 | 23 | 72 | {'max_depth': 23, 'min_samples_leaf': 72} | 0.849524 | 0.862857 | 0.862595 | ... | 0.854308 | 0.010440 | 7 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 1 | 0.002923 | 0.000063 | 0.000440 | 0.000072 | 26 | 83 | {'max_depth': 26, 'min_samples_leaf': 83} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 2 | 0.003855 | 0.000076 | 0.000390 | 0.000006 | 17 | 24 | {'max_depth': 17, 'min_samples_leaf': 24} | 0.847619 | 0.860952 | 0.868321 | ... | 0.854310 | 0.012162 | 6 | 0.875536 | 0.879351 | 0.878456 | 0.875596 | 0.881792 | 0.878146 | 0.002373 |
| 3 | 0.002825 | 0.000070 | 0.000420 | 0.000046 | 27 | 88 | {'max_depth': 27, 'min_samples_leaf': 88} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 4 | 0.002974 | 0.000045 | 0.000386 | 0.000021 | 31 | 64 | {'max_depth': 31, 'min_samples_leaf': 64} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.861296 | 0.860031 | 0.001460 |
| 5 | 0.002782 | 0.000039 | 0.000368 | 0.000006 | 27 | 89 | {'max_depth': 27, 'min_samples_leaf': 89} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.860343 | 0.859840 | 0.001340 |
| 6 | 0.003368 | 0.000051 | 0.000397 | 0.000019 | 22 | 42 | {'max_depth': 22, 'min_samples_leaf': 42} | 0.849524 | 0.836190 | 0.856870 | ... | 0.845540 | 0.015574 | 9 | 0.866476 | 0.865999 | 0.862726 | 0.865110 | 0.864156 | 0.864893 | 0.001343 |
| 7 | 0.003064 | 0.000083 | 0.000386 | 0.000011 | 21 | 62 | {'max_depth': 21, 'min_samples_leaf': 62} | 0.849524 | 0.840000 | 0.862595 | ... | 0.850500 | 0.009778 | 8 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.861296 | 0.860031 | 0.001460 |
| 8 | 0.002978 | 0.000048 | 0.000374 | 0.000008 | 20 | 64 | {'max_depth': 20, 'min_samples_leaf': 64} | 0.849524 | 0.862857 | 0.862595 | ... | 0.855072 | 0.009121 | 1 | 0.862184 | 0.858369 | 0.858913 | 0.859390 | 0.861296 | 0.860031 | 0.001460 |
9 rows × 22 columns
clf_RandomSearch = RandomSearch.best_estimator_
accuracy_RandomSearch = accuracy_score(y_test, clf_RandomSearch.predict(X_test))
print(f'Accuracy Manual: {accuracy_manual:.4f}')
print(f'Accuracy Grid search: {accuracy_GridSearch:.4f}')
print(f'Accuracy Random Search: {accuracy_RandomSearch:.4f}')
Accuracy Manual: 0.8201 Accuracy Grid search: 0.8430 Accuracy Random Search: 0.8430
Part D: Bayesian Optimization¶
- Expensive black box functions $\Rightarrow$ need of smart guesses
- Probabilistic Surrogate Model (to be fitted)
- Often Gaussian Processes
- Acquisition function
- Exploitation / Exploration
- Cheap to Computer
[Brochu, Cora, de Freitas, 2010]
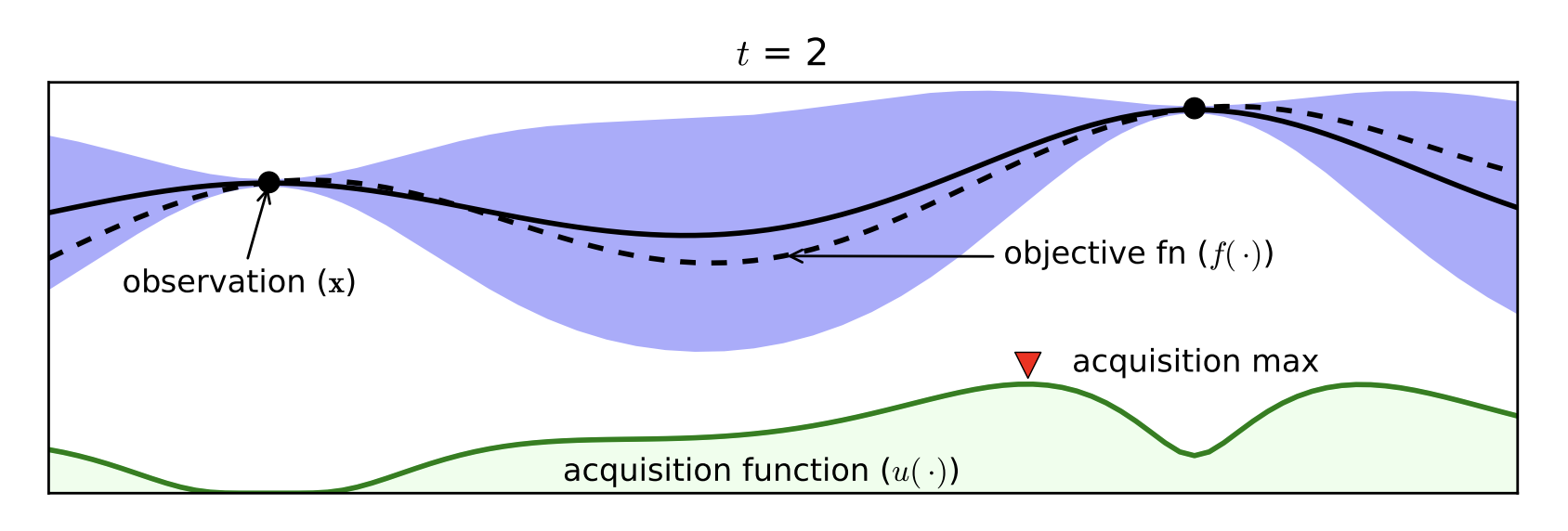
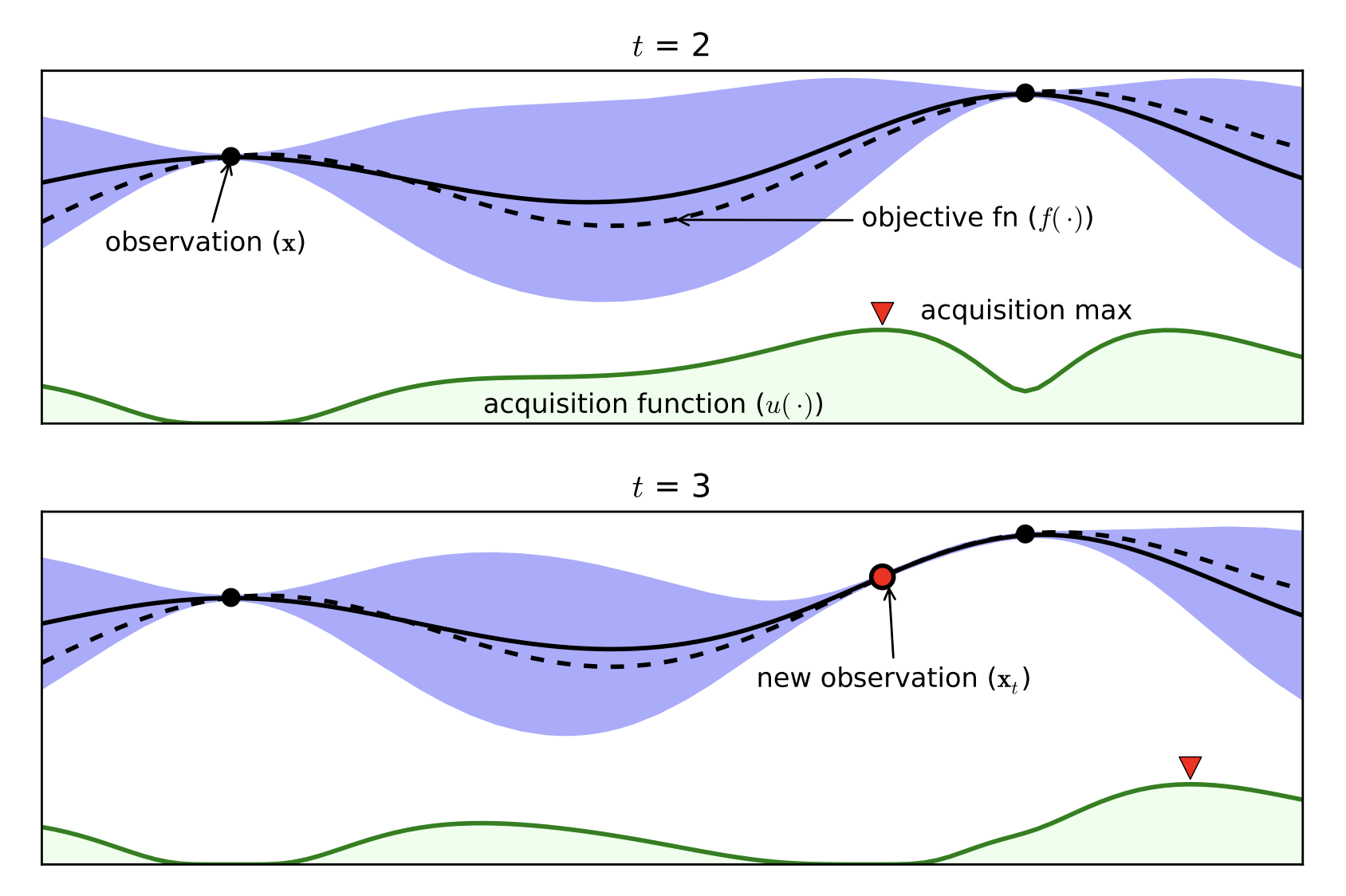
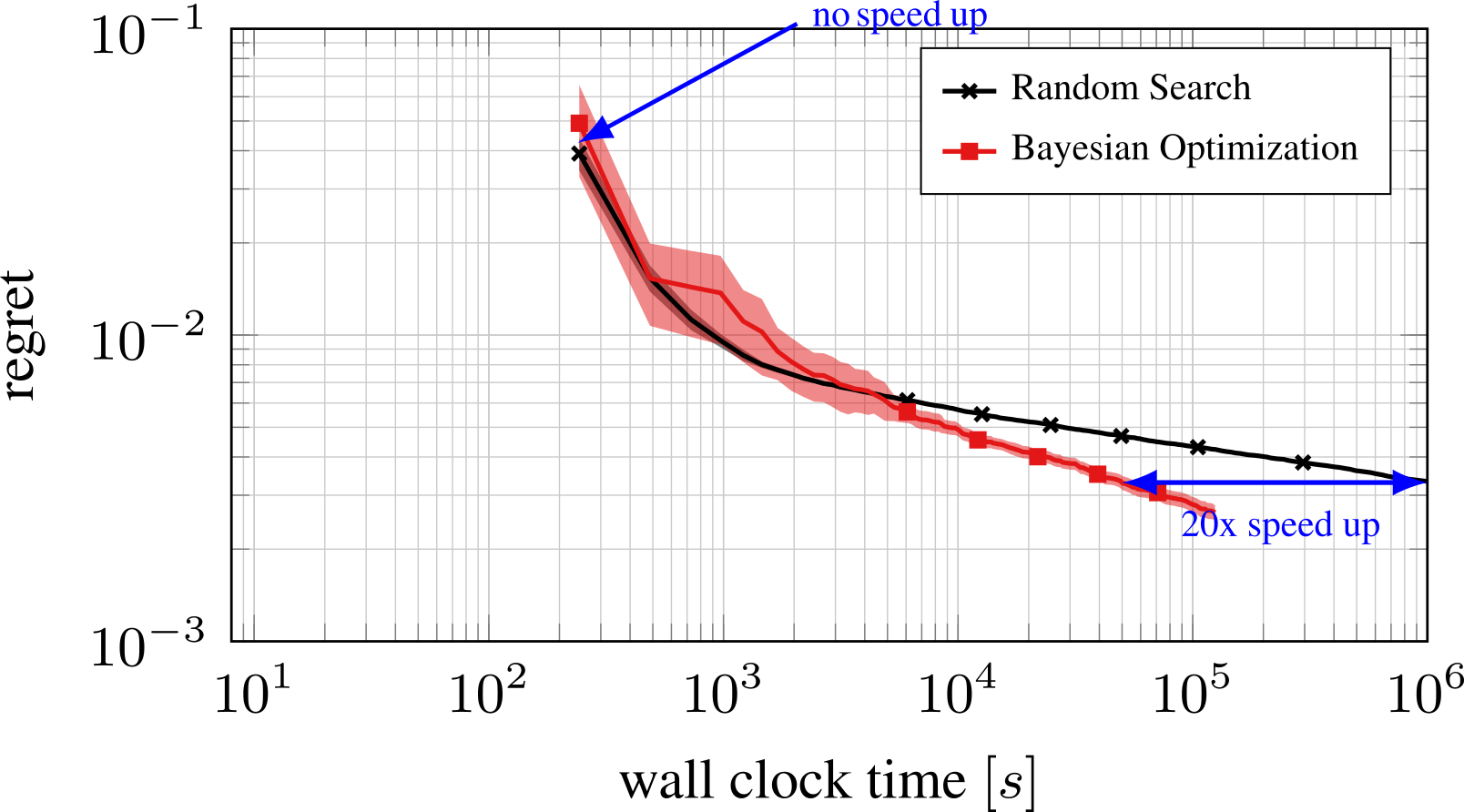
Bayesian Optimization with the Python package BayesianOptimization:
from bayes_opt import BayesianOptimization
from sklearn.model_selection import cross_val_score
def DecisionTree_CrossValidation(max_depth, min_samples_leaf, data, targets):
"""Decision Tree cross validation.
Fits a Decision Tree with the given paramaters to the target
given data, calculated a CV accuracy score and returns the mean.
The goal is to find combinations of max_depth, min_samples_leaf
that maximize the accuracy
"""
estimator = DecisionTreeClassifier(random_state=42,
max_depth=max_depth,
min_samples_leaf=min_samples_leaf)
cval = cross_val_score(estimator, data, targets, scoring='accuracy', cv=5)
return cval.mean()
def optimize_DecisionTree(data, targets, pars, n_iter=5):
"""Apply Bayesian Optimization to Decision Tree parameters."""
def crossval_wrapper(max_depth, min_samples_leaf):
"""Wrapper of Decision Tree cross validation.
Notice how we ensure max_depth, min_samples_leaf
are casted to integer before we pass them along.
"""
return DecisionTree_CrossValidation(max_depth=int(max_depth),
min_samples_leaf=int(min_samples_leaf),
data=data,
targets=targets)
optimizer = BayesianOptimization(f=crossval_wrapper,
pbounds=pars,
random_state=42,
verbose=2)
optimizer.maximize(init_points=4, n_iter=n_iter)
return optimizer
parameters_BayesianOptimization = {"max_depth": (1, 100),
"min_samples_leaf": (1, 100),
}
BayesianOptimization = optimize_DecisionTree(X_train,
y_train,
parameters_BayesianOptimization,
n_iter=5)
print(BayesianOptimization.max)
| iter | target | max_depth | min_sa... |
-------------------------------------------------
| 1 | 0.8551 | 38.08 | 95.12 |
| 2 | 0.849 | 73.47 | 60.27 |
| 3 | 0.8524 | 16.45 | 16.44 |
| 4 | 0.8551 | 6.75 | 86.75 |
| 5 | 0.8551 | 22.46 | 67.29 |
| 6 | 0.8551 | 21.86 | 65.88 |
| 7 | 0.8242 | 96.61 | 1.032 |
| 8 | 0.8551 | 98.91 | 99.99 |
| 9 | 0.8551 | 1.016 | 37.9 |
=================================================
{'target': np.float64(0.8550716103235187), 'params': {'max_depth': np.float64(38.07947176588889), 'min_samples_leaf': np.float64(95.1207163345817)}}
params = BayesianOptimization.max['params']
clf_BO = DecisionTreeClassifier(random_state=42, **params)
clf_BO = clf_BO.fit(X_train, y_train)
accuracy_BayesianOptimization = accuracy_score(y_test, clf_BO.predict(X_test))
print(f'Accuracy Manual: {accuracy_manual:.4f}')
print(f'Accuracy Grid Search: {accuracy_GridSearch:.4f}')
print(f'Accuracy Random Search: {accuracy_RandomSearch:.4f}')
print(f'Accuracy Bayesian Optimization: {accuracy_BayesianOptimization:.4f}')
Accuracy Manual: 0.8201 Accuracy Grid Search: 0.8430 Accuracy Random Search: 0.8430 Accuracy Bayesian Optimization: 0.8430
Part D: Full Scan over Parameter Space¶
Only possible in low-dimensional space, slow
max_depth_array = np.arange(1, 30)
min_samples_leaf_array = np.arange(2, 31)
Z = np.zeros((len(max_depth_array), len(min_samples_leaf_array)))
for i, max_depth in enumerate(max_depth_array):
for j, min_samples_leaf in enumerate(min_samples_leaf_array):
clf = DecisionTreeClassifier(random_state=42,
max_depth=max_depth,
min_samples_leaf=
min_samples_leaf)
clf.fit(X_train, y_train)
acc = accuracy_score(y_test, clf.predict(X_test))
Z[i, j] = acc
# Notice: have to transpose Z to match up with imshow
Z = Z.T
Plot the results:
fig, ax = plt.subplots(figsize=(12, 6))
# notice that we are setting the extent and origin keywords
CS = ax.imshow(Z, extent=[1, 30, 2, 31], cmap='viridis', origin='lower')
ax.set(xlabel='max_depth', ylabel='min_samples_leaf')
fig.colorbar(CS);
Sum up:¶
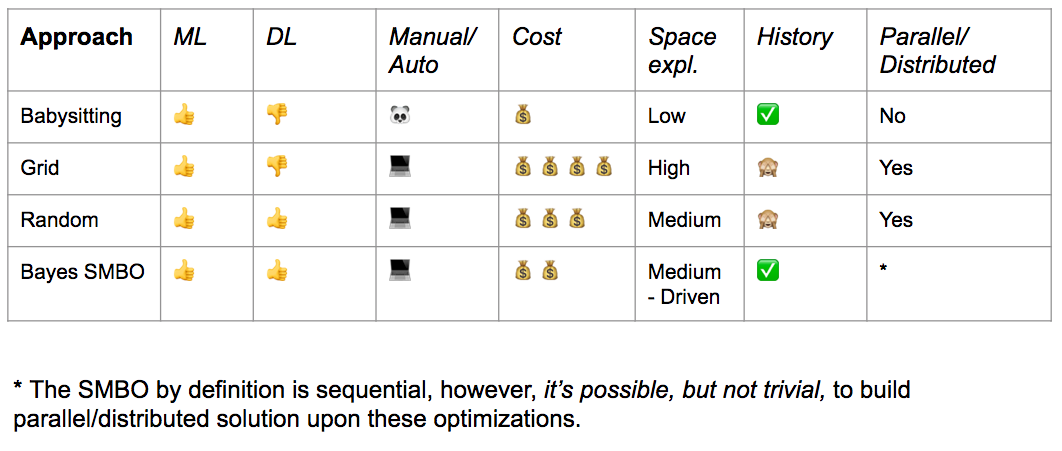
Part E: New Methods¶
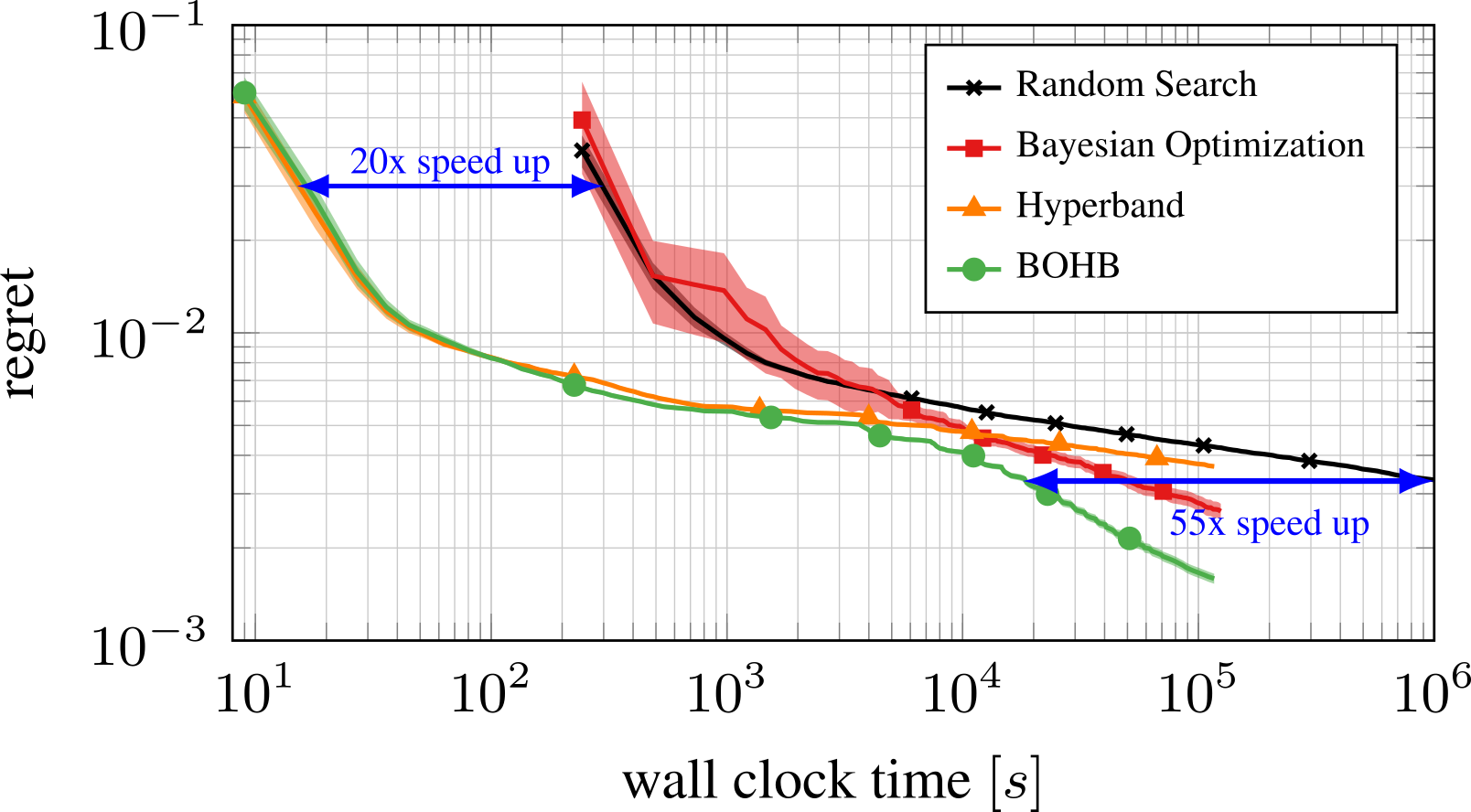
Bayesian Optimization meets HyperBand (BOHB)
HyperBand:
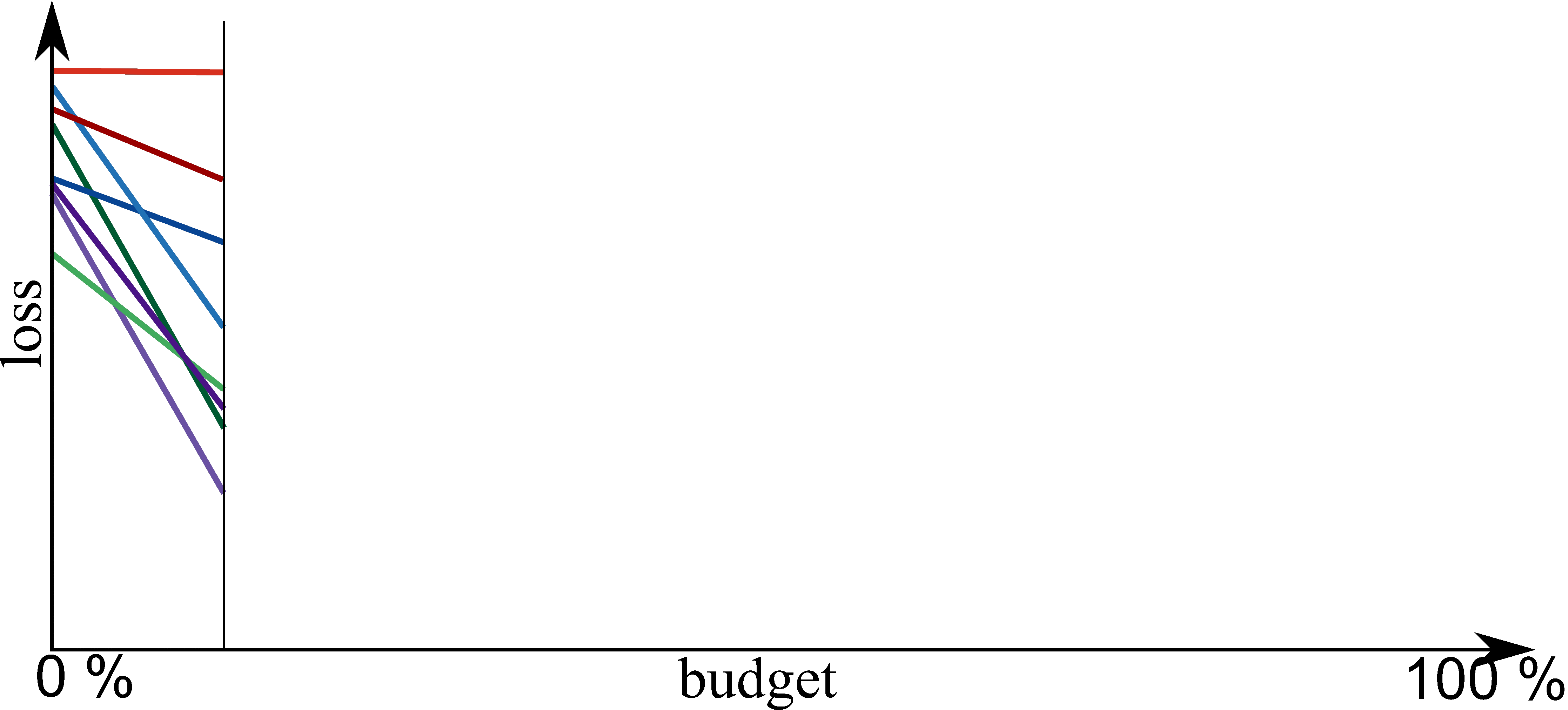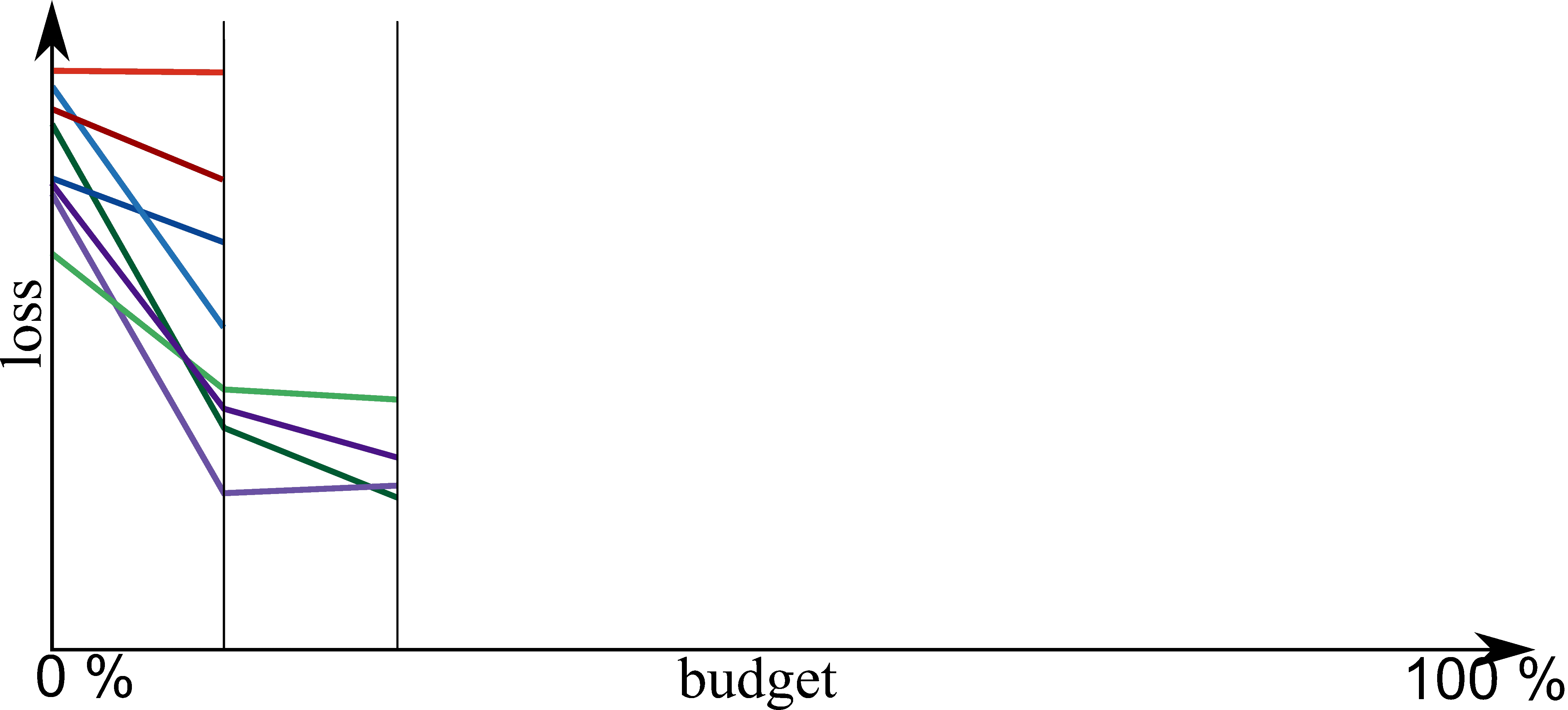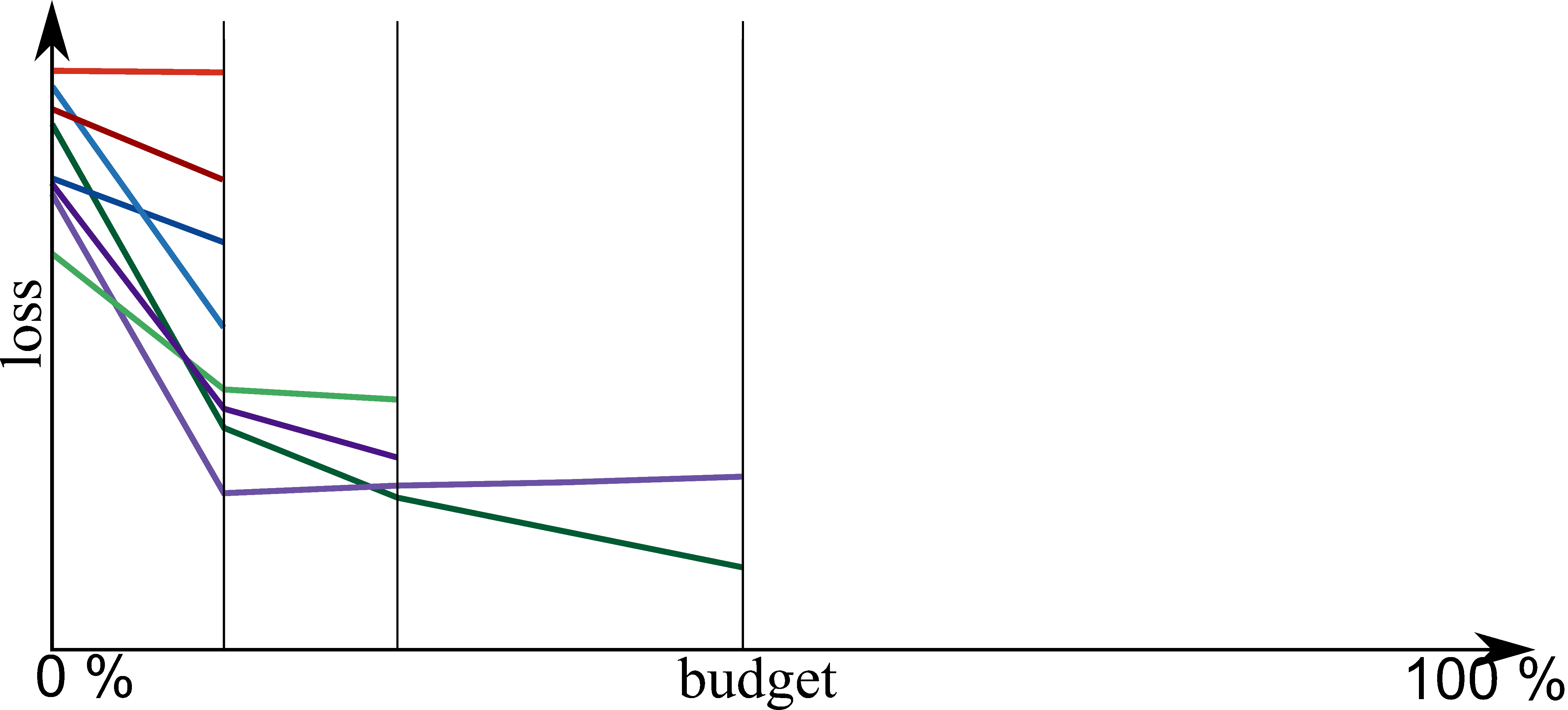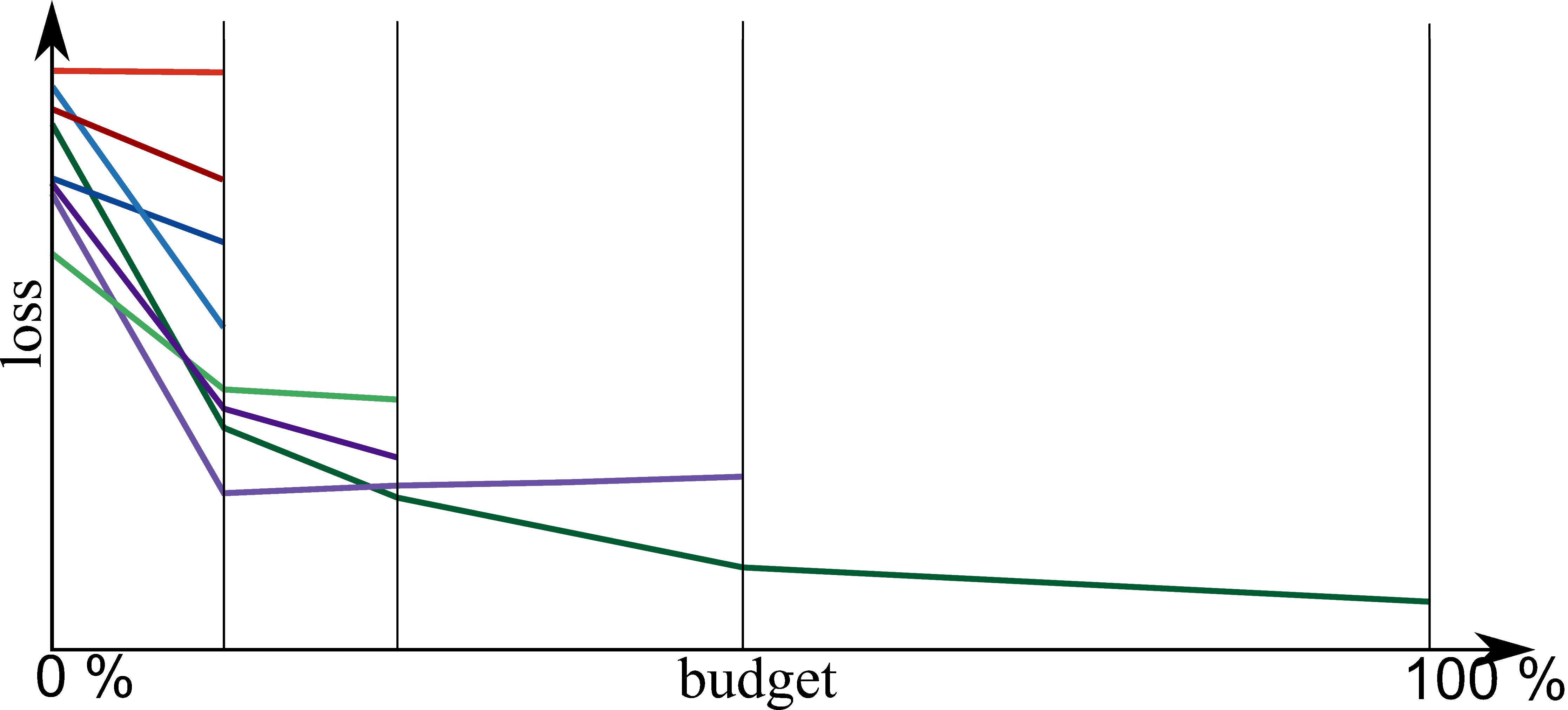

import optuna
from optuna.samplers import TPESampler
from optuna.integration import LightGBMPruningCallback
from optuna.pruners import MedianPruner
import lightgbm as lgb
lgb_data_train = lgb.Dataset(X_train, label=y_train);
def objective(trial):
boosting_types = ["gbdt", "rf", "dart"]
boosting_type = trial.suggest_categorical("boosting_type", boosting_types)
params = {
"objective": "binary",
"metric": 'auc',
"boosting": boosting_type,
"max_depth": 5,
"max_depth": trial.suggest_int("max_depth", 2, 63),
"min_child_weight": trial.suggest_float("min_child_weight", 1e-5, 10, log=True),
"scale_pos_weight": trial.suggest_float("scale_pos_weight", 10.0, 30.0),
"bagging_freq": 1, "bagging_fraction": 0.6,
"verbosity": -1
}
N_iterations_max = 10_000
early_stopping_rounds = 50
if boosting_type == "dart":
N_iterations_max = 100
early_stopping_rounds = 0
cv_res = lgb.cv(
params,
lgb_data_train,
num_boost_round=N_iterations_max,
seed=42,
callbacks=[LightGBMPruningCallback(trial, "auc"),lgb.early_stopping(stopping_rounds=early_stopping_rounds),lgb.log_evaluation(period=0)],
)
num_boost_round = len(cv_res["valid auc-mean"])
trial.set_user_attr("num_boost_round", num_boost_round)
return cv_res["valid auc-mean"][-1]
study = optuna.create_study(
direction="maximize",
sampler=TPESampler(seed=42),
pruner=MedianPruner(n_warmup_steps=50),
)
study.optimize(objective, n_trials=100, show_progress_bar=True);
[I 2025-04-28 09:31:18,530] A new study created in memory with name: no-name-1d1f7b80-84d6-44bb-b39d-3d860a4e34b7
0%| | 0/100 [00:00<?, ?it/s]
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[27] valid's auc: 0.923591 + 0.00625717
[I 2025-04-28 09:31:19,548] Trial 0 finished with value: 0.923591091502377 and parameters: {'boosting_type': 'rf', 'max_depth': 39, 'min_child_weight': 8.632008168602535e-05, 'scale_pos_weight': 13.119890406724053}. Best is trial 0 with value: 0.923591091502377.
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[30] valid's auc: 0.922688 + 0.00562831
[I 2025-04-28 09:31:20,491] Trial 1 finished with value: 0.9226879026588939 and parameters: {'boosting_type': 'rf', 'max_depth': 45, 'min_child_weight': 1.3289448722869181e-05, 'scale_pos_weight': 29.398197043239886}. Best is trial 0 with value: 0.923591091502377.
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[75] valid's auc: 0.931813 + 0.00638752
[I 2025-04-28 09:31:21,907] Trial 2 finished with value: 0.9318133275583781 and parameters: {'boosting_type': 'gbdt', 'max_depth': 13, 'min_child_weight': 0.0006690421166498799, 'scale_pos_weight': 20.495128632644757}. Best is trial 2 with value: 0.9318133275583781.
/opt/miniconda3/envs/appml/lib/python3.12/site-packages/lightgbm/callback.py:333: UserWarning: Early stopping is not available in dart mode
_log_warning("Early stopping is not available in dart mode")
[I 2025-04-28 09:31:23,399] Trial 3 finished with value: 0.9300085140349988 and parameters: {'boosting_type': 'dart', 'max_depth': 10, 'min_child_weight': 0.0005660670699258885, 'scale_pos_weight': 17.327236865873836}. Best is trial 2 with value: 0.9318133275583781.
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[23] valid's auc: 0.925103 + 0.00664956
[I 2025-04-28 09:31:24,162] Trial 4 finished with value: 0.925103266293118 and parameters: {'boosting_type': 'rf', 'max_depth': 33, 'min_child_weight': 0.035849855803404725, 'scale_pos_weight': 10.929008254399955}. Best is trial 2 with value: 0.9318133275583781.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:25,189] Trial 5 pruned. Trial was pruned at iteration 80.
/opt/miniconda3/envs/appml/lib/python3.12/site-packages/lightgbm/callback.py:333: UserWarning: Early stopping is not available in dart mode
_log_warning("Early stopping is not available in dart mode")
[I 2025-04-28 09:31:26,127] Trial 6 pruned. Trial was pruned at iteration 77. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:31:26,704] Trial 7 pruned. Trial was pruned at iteration 50.
/opt/miniconda3/envs/appml/lib/python3.12/site-packages/lightgbm/callback.py:333: UserWarning: Early stopping is not available in dart mode
_log_warning("Early stopping is not available in dart mode")
[I 2025-04-28 09:31:27,611] Trial 8 pruned. Trial was pruned at iteration 77.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:28,145] Trial 9 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:28,247] Trial 10 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[80] valid's auc: 0.932005 + 0.00572181
[I 2025-04-28 09:31:29,698] Trial 11 finished with value: 0.9320045589835481 and parameters: {'boosting_type': 'gbdt', 'max_depth': 17, 'min_child_weight': 0.0014474404511749194, 'scale_pos_weight': 17.256467103396812}. Best is trial 11 with value: 0.9320045589835481.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:30,722] Trial 12 pruned. Trial was pruned at iteration 91.
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[75] valid's auc: 0.932222 + 0.00647982
[I 2025-04-28 09:31:32,118] Trial 13 finished with value: 0.9322223804893104 and parameters: {'boosting_type': 'gbdt', 'max_depth': 21, 'min_child_weight': 0.009013452524358742, 'scale_pos_weight': 16.365725002496163}. Best is trial 13 with value: 0.9322223804893104.
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[80] valid's auc: 0.932962 + 0.00616164
[I 2025-04-28 09:31:33,593] Trial 14 finished with value: 0.9329620531129151 and parameters: {'boosting_type': 'gbdt', 'max_depth': 24, 'min_child_weight': 0.5206341066619701, 'scale_pos_weight': 14.999056028284222}. Best is trial 14 with value: 0.9329620531129151.
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[76] valid's auc: 0.932264 + 0.00804279
[I 2025-04-28 09:31:35,016] Trial 15 finished with value: 0.9322644588626462 and parameters: {'boosting_type': 'gbdt', 'max_depth': 27, 'min_child_weight': 0.4681880870979293, 'scale_pos_weight': 14.221884014764331}. Best is trial 14 with value: 0.9329620531129151.
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[76] valid's auc: 0.932096 + 0.00682524
[I 2025-04-28 09:31:36,380] Trial 16 finished with value: 0.9320961707531623 and parameters: {'boosting_type': 'gbdt', 'max_depth': 28, 'min_child_weight': 0.5699708687865693, 'scale_pos_weight': 13.290243259699489}. Best is trial 14 with value: 0.9329620531129151.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:37,232] Trial 17 pruned. Trial was pruned at iteration 73.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:37,856] Trial 18 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:38,085] Trial 19 pruned. Trial was pruned at iteration 50.
/opt/miniconda3/envs/appml/lib/python3.12/site-packages/lightgbm/callback.py:333: UserWarning: Early stopping is not available in dart mode
_log_warning("Early stopping is not available in dart mode")
[I 2025-04-28 09:31:38,740] Trial 20 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:39,319] Trial 21 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:40,244] Trial 22 pruned. Trial was pruned at iteration 65.
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[80] valid's auc: 0.932846 + 0.00661706
[I 2025-04-28 09:31:41,925] Trial 23 finished with value: 0.9328455584121311 and parameters: {'boosting_type': 'gbdt', 'max_depth': 38, 'min_child_weight': 0.13924551140538585, 'scale_pos_weight': 15.500812736247685}. Best is trial 14 with value: 0.9329620531129151.
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[78] valid's auc: 0.933695 + 0.00682342
[I 2025-04-28 09:31:43,338] Trial 24 finished with value: 0.933694616982525 and parameters: {'boosting_type': 'gbdt', 'max_depth': 38, 'min_child_weight': 0.17234934682104477, 'scale_pos_weight': 14.408730995874038}. Best is trial 24 with value: 0.933694616982525.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:44,063] Trial 25 pruned. Trial was pruned at iteration 60.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:44,667] Trial 26 pruned. Trial was pruned at iteration 58.
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[80] valid's auc: 0.93349 + 0.00699772
[I 2025-04-28 09:31:46,131] Trial 27 finished with value: 0.9334896829214714 and parameters: {'boosting_type': 'gbdt', 'max_depth': 53, 'min_child_weight': 0.1638624773314598, 'scale_pos_weight': 18.38632558260704}. Best is trial 24 with value: 0.933694616982525.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:46,755] Trial 28 pruned. Trial was pruned at iteration 50.
/opt/miniconda3/envs/appml/lib/python3.12/site-packages/lightgbm/callback.py:333: UserWarning: Early stopping is not available in dart mode
_log_warning("Early stopping is not available in dart mode")
[I 2025-04-28 09:31:47,397] Trial 29 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:48,060] Trial 30 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[80] valid's auc: 0.933762 + 0.00709228
[I 2025-04-28 09:31:49,520] Trial 31 finished with value: 0.9337617458077665 and parameters: {'boosting_type': 'gbdt', 'max_depth': 41, 'min_child_weight': 0.17576931740572696, 'scale_pos_weight': 18.394566552136062}. Best is trial 31 with value: 0.9337617458077665.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:50,091] Trial 32 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:50,665] Trial 33 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:51,257] Trial 34 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:51,808] Trial 35 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:52,342] Trial 36 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:52,924] Trial 37 pruned. Trial was pruned at iteration 51.
/opt/miniconda3/envs/appml/lib/python3.12/site-packages/lightgbm/callback.py:333: UserWarning: Early stopping is not available in dart mode
_log_warning("Early stopping is not available in dart mode")
[I 2025-04-28 09:31:53,486] Trial 38 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:31:54,045] Trial 39 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:31:54,570] Trial 40 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:31:55,200] Trial 41 pruned. Trial was pruned at iteration 57. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:31:56,089] Trial 42 pruned. Trial was pruned at iteration 73. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:31:56,655] Trial 43 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:31:57,251] Trial 44 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:31:57,810] Trial 45 pruned. Trial was pruned at iteration 51.
/opt/miniconda3/envs/appml/lib/python3.12/site-packages/lightgbm/callback.py:333: UserWarning: Early stopping is not available in dart mode
_log_warning("Early stopping is not available in dart mode")
[I 2025-04-28 09:31:58,391] Trial 46 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:58,943] Trial 47 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:31:59,451] Trial 48 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:32:00,034] Trial 49 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
Early stopping, best iteration is:
[76] valid's auc: 0.933483 + 0.00674034
[I 2025-04-28 09:32:01,373] Trial 50 finished with value: 0.9334828797925363 and parameters: {'boosting_type': 'gbdt', 'max_depth': 18, 'min_child_weight': 0.01738532870090808, 'scale_pos_weight': 13.694525540919285}. Best is trial 31 with value: 0.9337617458077665.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:32:01,933] Trial 51 pruned. Trial was pruned at iteration 51.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:32:02,476] Trial 52 pruned. Trial was pruned at iteration 58.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:32:03,017] Trial 53 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:32:03,564] Trial 54 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:32:04,092] Trial 55 pruned. Trial was pruned at iteration 50.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:32:04,921] Trial 56 pruned. Trial was pruned at iteration 74.
Training until validation scores don't improve for 50 rounds
[I 2025-04-28 09:32:05,563] Trial 57 pruned. Trial was pruned at iteration 60.
/opt/miniconda3/envs/appml/lib/python3.12/site-packages/lightgbm/callback.py:333: UserWarning: Early stopping is not available in dart mode
_log_warning("Early stopping is not available in dart mode")
[I 2025-04-28 09:32:06,236] Trial 58 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:06,808] Trial 59 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:07,461] Trial 60 pruned. Trial was pruned at iteration 61. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:08,097] Trial 61 pruned. Trial was pruned at iteration 59. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:08,761] Trial 62 pruned. Trial was pruned at iteration 60. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:09,412] Trial 63 pruned. Trial was pruned at iteration 51. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:09,993] Trial 64 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:10,592] Trial 65 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:11,164] Trial 66 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:11,710] Trial 67 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:12,276] Trial 68 pruned. Trial was pruned at iteration 51. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:13,086] Trial 69 pruned. Trial was pruned at iteration 73.
/opt/miniconda3/envs/appml/lib/python3.12/site-packages/lightgbm/callback.py:333: UserWarning: Early stopping is not available in dart mode
_log_warning("Early stopping is not available in dart mode")
[I 2025-04-28 09:32:13,682] Trial 70 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:14,290] Trial 71 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:14,832] Trial 72 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:15,365] Trial 73 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:15,908] Trial 74 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:16,457] Trial 75 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:17,170] Trial 76 pruned. Trial was pruned at iteration 51. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:18,086] Trial 77 pruned. Trial was pruned at iteration 74. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:18,724] Trial 78 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:19,290] Trial 79 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:19,852] Trial 80 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:20,999] Trial 81 pruned. Trial was pruned at iteration 60. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:21,670] Trial 82 pruned. Trial was pruned at iteration 60. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:22,575] Trial 83 pruned. Trial was pruned at iteration 60. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:23,162] Trial 84 pruned. Trial was pruned at iteration 51. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:23,713] Trial 85 pruned. Trial was pruned at iteration 50.
/opt/miniconda3/envs/appml/lib/python3.12/site-packages/lightgbm/callback.py:333: UserWarning: Early stopping is not available in dart mode
_log_warning("Early stopping is not available in dart mode")
[I 2025-04-28 09:32:24,305] Trial 86 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:24,844] Trial 87 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:25,413] Trial 88 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:25,999] Trial 89 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:26,541] Trial 90 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:27,095] Trial 91 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:27,671] Trial 92 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:28,227] Trial 93 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:28,863] Trial 94 pruned. Trial was pruned at iteration 61. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:29,453] Trial 95 pruned. Trial was pruned at iteration 50. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:30,067] Trial 96 pruned. Trial was pruned at iteration 51. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:30,726] Trial 97 pruned. Trial was pruned at iteration 60. Training until validation scores don't improve for 50 rounds [I 2025-04-28 09:32:31,476] Trial 98 pruned. Trial was pruned at iteration 61.
/opt/miniconda3/envs/appml/lib/python3.12/site-packages/lightgbm/callback.py:333: UserWarning: Early stopping is not available in dart mode
_log_warning("Early stopping is not available in dart mode")
[I 2025-04-28 09:32:32,063] Trial 99 pruned. Trial was pruned at iteration 50.
# To see all info at the best trial use:
study.best_trial
# To print metric values for all trials:
study.best_trial.intermediate_values
# To see distributions from which optuna samples parameters:
study.best_trial.distributions
{'boosting_type': CategoricalDistribution(choices=('gbdt', 'rf', 'dart')),
'max_depth': IntDistribution(high=63, log=False, low=2, step=1),
'min_child_weight': FloatDistribution(high=10.0, log=True, low=1e-05, step=None),
'scale_pos_weight': FloatDistribution(high=30.0, log=False, low=10.0, step=None)}
# To simply get the optimized parameters:
study.best_trial.params
{'boosting_type': 'gbdt',
'max_depth': 41,
'min_child_weight': 0.17576931740572696,
'scale_pos_weight': 18.394566552136062}
Happy HyperParameter Optimisation!¶
...and remember, that this is useful but not essential in this course.